Reporter Flaviviruses as Tools to Demonstrate Homologous and Heterologous Superinfection Exclusion
- PMID: 35891480
- PMCID: PMC9317482
- DOI: 10.3390/v14071501
Reporter Flaviviruses as Tools to Demonstrate Homologous and Heterologous Superinfection Exclusion
Abstract
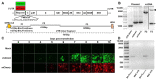
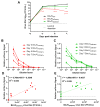
Binjari virus (BinJV) is a lineage II or dual-host affiliated insect-specific flavivirus previously demonstrated as replication-deficient in vertebrate cells. Previous studies have shown that BinJV is tolerant to exchanging its structural proteins (prM-E) with pathogenic flaviviruses, making it a safe backbone for flavivirus vaccines. Here, we report generation by circular polymerase extension reaction of BinJV expressing zsGreen or mCherry fluorescent protein. Recovered BinJV reporter viruses grew to high titres (107-8 FFU/mL) in Aedes albopictus C6/36 cells assayed using immunoplaque assays (iPA). We also demonstrate that BinJV reporters could be semi-quantified live in vitro using a fluorescence microplate reader with an observed linear correlation between quantified fluorescence of BinJV reporter virus-infected C6/36 cells and iPA-quantitated virus titres. The utility of the BinJV reporter viruses was then examined in homologous and heterologous superinfection exclusion assays. We demonstrate that primary infection of C6/36 cells with BinJVzsGreen completely inhibits a secondary infection with homologous BinJVmCherry or heterologous ZIKVmCherry using fluorescence microscopy and virus quantitation by iPA. Finally, BinJVzsGreen infections were examined in vivo by microinjection of Aedes aegypti with BinJVzsGreen. At seven days post-infection, a strong fluorescence in the vicinity of salivary glands was detected in frozen sections. This is the first report on the construction of reporter viruses for lineage II insect-specific flaviviruses and establishes a tractable system for exploring flavivirus superinfection exclusion in vitro and in vivo.
Keywords: Binjari virus; CPER; flavivirus; insect-specific viruses; reporter viruses; superinfection exclusion.
Conflict of interest statement
N.D.N., L.J.V., J.H.-P., R.A.H. and A.A.K. are inventors on a patent (WO/2018/176075) covering the Binjari chimera technology. No other authors declare competing interests.
Figures



References
-
- Stanaway J.D., Shepard D.S., Undurraga E.A., Halasa Y.A., Coffeng L.E., Brady O.J., Hay S.I., Bedi N., Bensenor I.M., Castaneda-Orjuela C.A., et al. The global burden of dengue: An analysis from the Global Burden of Disease Study 2013. Lancet Infect. Dis. 2016;16:712–723. doi: 10.1016/S1473-3099(16)00026-8. - DOI - PMC - PubMed
-
- Roby J.A., Funk A., Khromykh A.A. Flavivirus replication and assembly. In: Shi P.Y., editor. Molecular Virology and Control of Flaviviruses. Caister Academic Press; Norfolk, UK: 2012. pp. 21–49.
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical

