Nipah Virus Infection Generates Ordered Structures in Cellulo
- PMID: 35891503
- PMCID: PMC9317923
- DOI: 10.3390/v14071523
Nipah Virus Infection Generates Ordered Structures in Cellulo
Abstract
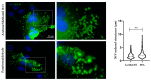
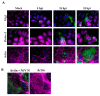
Nipah virus (NiV) is a zoonotic paramyxovirus with a fatality rate of up to 92% in humans. While several pathogenic mechanisms used by NiV to counteract host immune defense responses have been described, all of the processes that take place in cells during infection are not fully characterized. Here, we describe the formation of ordered intracellular structures during NiV infection. We observed that these structures are formed specifically during NiV infection, but not with other viruses from the same Mononegavirales order (namely Ebola virus) or from other orders such as Bunyavirales (Junín virus). We also determined the kinetics of the appearance of these structures and their cellular localization at the cellular periphery. Finally, we confirmed the presence of these NiV-specific ordered structures using structured illumination microscopy (SIM), as well as their localization by transmission electron microscopy (TEM), scanning electron microscopy (SEM), and correlative light and electron microscopy (CLEM). Herein, we describe a cytopathogenic mechanism that provides a new insight into NiV biology. These newly described ordered structures could provide a target for novel antiviral approaches.
Keywords: CLEM; Nipah virus; SEM; SIM; TEM; in cellulo; ordered structures.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

