Self-Repairing Herpesvirus Saimiri Deletion Variants
- PMID: 35891505
- PMCID: PMC9320899
- DOI: 10.3390/v14071525
Self-Repairing Herpesvirus Saimiri Deletion Variants
Abstract
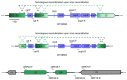
Herpesvirus saimiri (HVS) is discussed as a possible vector in gene therapy. In order to create a self-repairing HVS vector, the F plasmid vector moiety of the bacterial artificial chromosome (BAC) was transposed via Red recombination into the virus genes ORF22 or ORF29b, both important for virus replication. Repetitive sequences were additionally inserted, allowing the removal of the F-derived sequences from the viral DNA genome upon reconstitution in permissive epithelial cells. Moreover, these self-repair-enabled BACs were used to generate deletion variants of the transforming strain C488 in order to minimalize the virus genome. Using the en passant mutagenesis with two subsequent homologous recombination steps, the BAC was seamlessly manipulated. To ensure the replication capacity in permissive monkey cells, replication kinetics for all generated virus variants were documented. HVS variants with increased insert capacity reached the self-repair within two to three passages in permissive epithelial cells. The seamless deletion of ORFs 3/21, 12-14, 16 or 71 did not abolish replication competence. Apoptosis induction did not seem to be altered in human T cells transformed with deletion variants lacking ORF16 or ORF71. These virus variants form an important step towards creating a potential minimal virus vector for gene therapy, for example, in human T cells.
Keywords: herpesvirus saimiri; recombination; vector.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Generation and precise modification of a herpesvirus saimiri bacterial artificial chromosome demonstrates that the terminal repeats are required for both virus production and episomal persistence.J Gen Virol. 2003 Dec;84(Pt 12):3393-3403. doi: 10.1099/vir.0.19387-0. J Gen Virol. 2003. PMID: 14645920
-
Red-mediated transposition and final release of the mini-F vector of a cloned infectious herpesvirus genome.PLoS One. 2009 Dec 4;4(12):e8178. doi: 10.1371/journal.pone.0008178. PLoS One. 2009. PMID: 19997639 Free PMC article.
-
Herpesvirus saimiri.Philos Trans R Soc Lond B Biol Sci. 2001 Apr 29;356(1408):545-67. doi: 10.1098/rstb.2000.0780. Philos Trans R Soc Lond B Biol Sci. 2001. PMID: 11313011 Free PMC article. Review.
-
Production of recombinant Herpesvirus saimiri-based vectors.Cold Spring Harb Protoc. 2011 Dec 1;2011(12):1515-9. doi: 10.1101/pdb.prot066944. Cold Spring Harb Protoc. 2011. PMID: 22135662
-
Bovine herpesvirus 4: genomic organization and relationship with two other gammaherpesviruses, Epstein-Barr virus and herpesvirus saimiri.Vet Microbiol. 1996 Nov;53(1-2):79-89. doi: 10.1016/s0378-1135(96)01236-9. Vet Microbiol. 1996. PMID: 9011000 Review.
Cited by
-
Bacterial-Artificial-Chromosome-Based Genome Editing Methods and the Applications in Herpesvirus Research.Microorganisms. 2023 Feb 26;11(3):589. doi: 10.3390/microorganisms11030589. Microorganisms. 2023. PMID: 36985163 Free PMC article. Review.
References
-
- Meléndez L.V., Hunt R.D., Daniel M.D., García F.G., Fraser C.E. Herpesvirus Saimiri. II. Experimentally Induced Malignant Lymphoma in Primates. Lab. Anim. Care. 1969;19:378–386. - PubMed
-
- Hunt R.D., Meléndez L.V. Herpesvirus Infections of Non-human Primates: A Review. Lab. Anim. Care. 1969;19:221–234. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

