Origins and Evolution of Seasonal Human Coronaviruses
- PMID: 35891531
- PMCID: PMC9320361
- DOI: 10.3390/v14071551
Origins and Evolution of Seasonal Human Coronaviruses
Abstract
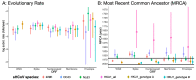
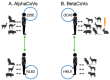
Four seasonal human coronaviruses (sHCoVs) are endemic globally (229E, NL63, OC43, and HKU1), accounting for 5-30% of human respiratory infections. However, the epidemiology and evolution of these CoVs remain understudied due to their association with mild symptomatology. Using a multigene and complete genome analysis approach, we find the evolutionary histories of sHCoVs to be highly complex, owing to frequent recombination of CoVs including within and between sHCoVs, and uncertain, due to the under sampling of non-human viruses. The recombination rate was highest for 229E and OC43 whereas substitutions per recombination event were highest in NL63 and HKU1. Depending on the gene studied, OC43 may have ungulate, canine, or rabbit CoV ancestors. 229E may have origins in a bat, camel, or an unsampled intermediate host. HKU1 had the earliest common ancestor (1809-1899) but fell into two distinct clades (genotypes A and B), possibly representing two independent transmission events from murine-origin CoVs that appear to be a single introduction due to large gaps in the sampling of CoVs in animals. In fact, genotype B was genetically more diverse than all the other sHCoVs. Finally, we found shared amino acid substitutions in multiple proteins along the non-human to sHCoV host-jump branches. The complex evolution of CoVs and their frequent host switches could benefit from continued surveillance of CoVs across non-human hosts.
Keywords: 229E; HKU1; NL63; OC43; evolution; recombination; seasonal coronaviruses; zoonosis.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

