Hemagglutinin Subtype Specificity and Mechanisms of Highly Pathogenic Avian Influenza Virus Genesis
- PMID: 35891546
- PMCID: PMC9321182
- DOI: 10.3390/v14071566
Hemagglutinin Subtype Specificity and Mechanisms of Highly Pathogenic Avian Influenza Virus Genesis
Abstract
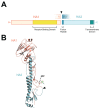
Highly Pathogenic Avian Influenza Viruses (HPAIVs) arise from low pathogenic precursors following spillover from wild waterfowl into poultry populations. The main virulence determinant of HPAIVs is the presence of a multi-basic cleavage site (MBCS) in the hemagglutinin (HA) glycoprotein. The MBCS allows for HA cleavage and, consequently, activation by ubiquitous proteases, which results in systemic dissemination in terrestrial poultry. Since 1959, 51 independent MBCS acquisition events have been documented, virtually all in HA from the H5 and H7 subtypes. In the present article, data from natural LPAIV to HPAIV conversions and experimental in vitro and in vivo studies were reviewed in order to compile recent advances in understanding HA cleavage efficiency, protease usage, and MBCS acquisition mechanisms. Finally, recent hypotheses that might explain the unique predisposition of the H5 and H7 HA sequences to obtain an MBCS in nature are discussed.
Keywords: Highly Pathogenic Avian Influenza Viruses; Low-Pathogenic Avian Influenza Viruses; RNA-dependent RNA polymerase; multibasic cleavage site; pathogen evolution; proteolytic cleavage; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Daoust P.Y., Kibenge F.S.B., Fouchier R.A.M., van de Bildt M.W.G., Van Riel D., Kuiken T. Replication of low pathogenic avian influenza virus in naturally infected mallard ducks (Anas platyrhynchos) causes no morphologic lesions. J. Wildl. Dis. 2011;47:401–409. doi: 10.7589/0090-3558-47.2.401. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

