Identification of a Virulent Newcastle Disease Virus Strain Isolated from Pigeons (Columbia livia) in Northeastern Brazil Using Next-Generation Genome Sequencing
- PMID: 35891559
- PMCID: PMC9319777
- DOI: 10.3390/v14071579
Identification of a Virulent Newcastle Disease Virus Strain Isolated from Pigeons (Columbia livia) in Northeastern Brazil Using Next-Generation Genome Sequencing
Abstract
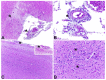
Newcastle disease virus (NDV), also known as avian paramyxoviruses 1 (APMV-1) is among the most important viruses infecting avian species. Given its widespread circulation, there is a high risk for the reintroduction of virulent strains into the domestic poultry industry, making the surveillance of wild and domestic birds a crucial process to appropriately respond to novel outbreaks. In the present study, we investigated an outbreak characterized by the identification of sick pigeons in a large municipality in Northeastern Brazil in 2018. The affected pigeons presented neurological signs, including motor incoordination, torticollis, and lethargy. Moribund birds were collected, and through a detailed histopathological analysis we identified severe lymphoplasmacytic meningoencephalitis with perivascular cuffs and gliosis in the central nervous system, and lymphoplasmacytic inflammation in the liver, kidney, and intestine. A total of five pigeons tested positive for NDV, as assessed by rRT-PCR targeted to the M gene. Laboratory virus isolation on Vero E6 cells confirmed infection, after the recovery of infectious NVD from brain and kidney tissues. We next characterized the isolated NDV/pigeon/PE-Brazil/MP003/2018 by next-generation sequencing (NGS). Phylogenetic analysis grouped the virus with other NDV class II isolates from subgenotype VI.2.1.2, including two previous NDV isolates from Brazil in 2014 and 2019. The diversity of aminoacid residues at the fusion F protein cleavage site was analyzed identifying the motif RRQKR↓F, typical of virulent strains. Our results all highlight the importance of virus surveillance in wild and domestic birds, especially given the risk of zoonotic NDV.
Keywords: Newcastle disease virus; avian paramyxovirus 1; domestic birds; pigeons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

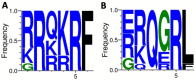
References
-
- Dimitrov K.M., Abolnik C., Afonso C.L., Albina E., Bahl J., Berg M., Briand F.X., Brown I.H., Choi K.S., Chvala I., et al. Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect. Genet. Evol. 2019;74:103917. doi: 10.1016/j.meegid.2019.103917. - DOI - PMC - PubMed
-
- Diel D.G., da Silva L.H., Liu H., Wang Z., Miller P.J., Afonso C.L. Genetic diversity of avian paramyxovirus type 1: Proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect. Genet. Evol. 2012;12:1770–1779. doi: 10.1016/j.meegid.2012.07.012. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

