Influenza A Virus Infection Reactivates Human Endogenous Retroviruses Associated with Modulation of Antiviral Immunity
- PMID: 35891571
- PMCID: PMC9320126
- DOI: 10.3390/v14071591
Influenza A Virus Infection Reactivates Human Endogenous Retroviruses Associated with Modulation of Antiviral Immunity
Abstract
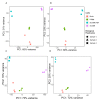
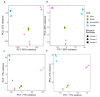
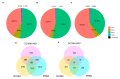
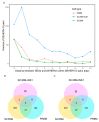
Human endogenous retrovirus (HERVs), normally silenced by methylation or mutations, can be reactivated by multiple environmental factors, including infections with exogenous viruses. In this work, we investigated the transcriptional activity of HERVs in human A549 cells infected by two wild-type (PR8M, SC35M) and one mutated (SC35MΔNS1) strains of Influenza A virus (IAVs). We found that the majority of differentially expressed HERVs (DEHERVS) and genes (DEGs) were up-regulated in the infected cells, with the most significantly enriched biological processes associated with the genes differentially expressed exclusively in SC35MΔNS1 being linked to the immune system. Most DEHERVs in PR8M and SC35M are mammalian apparent LTR retrotransposons, while in SC35MΔNS1, more HERV loci from the HERVW9 group were differentially expressed. Furthermore, up-regulated pairs of HERVs and genes in close chromosomal proximity to each other tended to be associated with immune responses, which implies that specific HERV groups might have the potential to trigger specific gene networks and influence host immunological pathways.
Keywords: expression analysis; functional annotation; gene regulation; genome analysis; immune response; virus-host interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Badarinarayan S.S., Shcherbakova I., Langer S., Koepke L., Preising A., Hotter D., Kirchhoff F., Sparrer K.M.J., Schotta G., Sauter D. HIV-1 infection activates endogenous retroviral promoters regulating antiviral gene expression. Nucleic Acids Res. 2020;48:10890–10908. doi: 10.1093/nar/gkaa832. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

