Characteristics of gut microbiota in representative mice strains: Implications for biological research
- PMID: 35892142
- PMCID: PMC9434578
- DOI: 10.1002/ame2.12257
Characteristics of gut microbiota in representative mice strains: Implications for biological research
Abstract
Background: Experimental animals are used to study physiological phenomena, pathological mechanisms, and disease prevention. The gut microbiome is known as a potential confounding factor for inconsistent data from preclinical studies. Although many gut microbiome studies have been conducted in recent decades, few have focused on gut microbiota fluctuation among representative mouse strains.
Methods: A range of frequently used mouse strains were selected from 34 isolation packages representing disease-related animal (DRA), immunity defect animal (IDA), or gene-editing animal (GEA) from the BALB/c and C57BL/6J backgrounds together with normal mice, and their microbial genomic DNA were isolated from mouse feces to sequence for the exploration of gut microbiota.
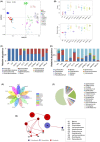
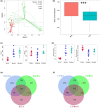
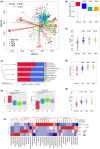
Results: Mouse background strain, classification, introduced source, introduced year, and reproduction type significantly affected the gut microbiota structure (p < 0.001 for all parameters), with background strain contributing the greatest influence (R2 = 0.237). In normal groups, distinct gut microbiota types existed in different mouse strains. Sixty-four core operational taxonomic units were obtained from normal mice, and 12 belonged to Lactobacillus. Interestingly, the gut microbiota in C57BL/6J was more stable than that in BALB/c mice. Furthermore, the gut microbiota in the IDA, GEA, and DRA groups significantly differed from that in normal groups (p < 0.001 for all). Compared with the normal group, there was a significantly higher Chao1 and Shannon index (p < 0.001 for all) in the IDA, GEA, and DRA groups. Markedly changed classes occurred with Firmicutes and Bacteroidetes. The abundances of Helicobacter, Blautia, Enterobacter, Bacillus, Clostridioides, Paenibacillus, and Clostridiales all significantly decreased in the IDA, GEA, and DRA groups, whereas those of Saccharimonas, Rikenella, and Odoribacter all significantly increased.
Keywords: Bacteroidetes; Firmicutes; BALB/c mice; C57BL/6J; disease-related animal; gene-editing animal; gut microbiota; immunity defect animal; strains.
© 2022 The Authors. Animal Models and Experimental Medicine published by John Wiley & Sons Australia, Ltd on behalf of The Chinese Association for Laboratory Animal Sciences.
Conflict of interest statement
The authors declare that they have no conflicts of interest. Jianguo Guo and Chuan Qin are editorial board members of Animal Models and Experimental Medicine and a coauthors of this article. To minimize bias, they were excluded from all editorial decision making related to the acceptance of this article for publication.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

