Integrative Organelle-Based Functional Proteomics: In Silico Prediction of Impaired Functional Annotations in SACS KO Cell Model
- PMID: 35892334
- PMCID: PMC9331974
- DOI: 10.3390/biom12081024
Integrative Organelle-Based Functional Proteomics: In Silico Prediction of Impaired Functional Annotations in SACS KO Cell Model
Abstract
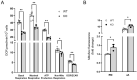
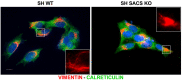
Autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS) is an inherited neurodegenerative disease characterized by early-onset spasticity in the lower limbs, axonal-demyelinating sensorimotor peripheral neuropathy, and cerebellar ataxia. Our understanding of ARSACS (genetic basis, protein function, and disease mechanisms) remains partial. The integrative use of organelle-based quantitative proteomics and whole-genome analysis proposed in the present study allowed identifying the affected disease-specific pathways, upstream regulators, and biological functions related to ARSACS, which exemplify a rationale for the development of improved early diagnostic strategies and alternative treatment options in this rare condition that currently lacks a cure. Our integrated results strengthen the evidence for disease-specific defects related to bioenergetics and protein quality control systems and reinforce the role of dysregulated cytoskeletal organization in the pathogenesis of ARSACS.
Keywords: ARSACS; KO models; SACS; biomarkers; lysosomes; mitochondria; omics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS) - A Polish family with novel SACS mutations.Neurol Neurochir Pol. 2017 Nov-Dec;51(6):481-485. doi: 10.1016/j.pjnns.2017.08.003. Epub 2017 Aug 17. Neurol Neurochir Pol. 2017. PMID: 28843771
-
A novel SACS p.Pro4154GlnfsTer20 mutation in a family with autosomal recessive spastic ataxia of Charlevoix-Saguenay.Neurol Sci. 2021 Jul;42(7):2969-2973. doi: 10.1007/s10072-021-05117-1. Epub 2021 Feb 9. Neurol Sci. 2021. PMID: 33559790
-
Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay without Spasticity.Intern Med. 2021 Dec 15;60(24):3963-3967. doi: 10.2169/internalmedicine.7401-21. Epub 2021 Jun 12. Intern Med. 2021. PMID: 34121011 Free PMC article.
-
Insights into SACS pathological attributes in autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS)☆.Curr Opin Chem Biol. 2022 Dec;71:102211. doi: 10.1016/j.cbpa.2022.102211. Epub 2022 Sep 17. Curr Opin Chem Biol. 2022. PMID: 36126381 Review.
-
Genetics of Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay (ARSACS) and Role of Sacsin in Neurodegeneration.Int J Mol Sci. 2022 Jan 4;23(1):552. doi: 10.3390/ijms23010552. Int J Mol Sci. 2022. PMID: 35008978 Free PMC article. Review.
Cited by
-
Long-term benefits of TUDCA supplement in ARSACS zebrafish model.Sci Rep. 2025 Jul 14;15(1):25429. doi: 10.1038/s41598-025-10850-0. Sci Rep. 2025. PMID: 40659819 Free PMC article.
-
Proteomics and lipidomic analysis reveal dysregulated pathways associated with loss of sacsin.Front Neurosci. 2024 Jun 7;18:1375299. doi: 10.3389/fnins.2024.1375299. eCollection 2024. Front Neurosci. 2024. PMID: 38911600 Free PMC article.
-
AlphaFold predicted structure of the Hsp90-like domains of the neurodegeneration linked protein sacsin reveals key residues for ATPase activity.Front Mol Biosci. 2023 Jan 13;9:1074714. doi: 10.3389/fmolb.2022.1074714. eCollection 2022. Front Mol Biosci. 2023. PMID: 36710881 Free PMC article.
-
Selection of Clinical Outcome Assessments for Trial Readiness in ARSACS - 2-year Progression and Responsiveness to Change Part 1: Disease Severity, Swallowing, Upper Limb Function, and Participation.Cerebellum. 2025 May 31;24(4):106. doi: 10.1007/s12311-025-01858-3. Cerebellum. 2025. PMID: 40450178
References
-
- Engert J.C., Bérubé P., Mercier J., Doré C., Lepage P., Ge B., Bouchard J.-P., Mathieu J., Melançon S.B., Schalling M., et al. ARSACS, a spastic ataxia common in northeastern Québec, is caused by mutations in a new gene encoding an 11.5-kb ORF. Nat. Genet. 2000;24:120–125. doi: 10.1038/72769. - DOI - PubMed
-
- Romano A., Tessa A., Barca A., Fattori F., de Leva M.F., Terracciano A., Storelli C., Santorelli F.M., Verri T. Comparative analysis and functional mapping of SACS mutations reveal novel insights into sacsin repeated architecture. Hum. Mutat. 2013;34:525–537. doi: 10.1002/humu.22269. - DOI - PMC - PubMed
-
- Girard M., Lariviere R., Parfitt D.A., Deane E.C., Gaudet R., Nossova N., Blondeau F., Prenosil G., Vermeulen E.G.M., Duchen M.R., et al. Mitochondrial dysfunction and Purkinje cell loss in autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS) Proc. Natl. Acad. Sci. USA. 2012;109:1661–1666. doi: 10.1073/pnas.1113166109. - DOI - PMC - PubMed
-
- Criscuolo C., Procaccini C., Meschini M.C., Cianflone A., Carbone R., Doccini S., Devos D., Nesti C., Vuillaume I., Pellegrino M., et al. Powerhouse failure and oxidative damage in autosomal recessive spastic ataxia of Charlevoix-Saguenay. J. Neurol. 2015;262:2755–2763. doi: 10.1007/s00415-015-7911-4. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials

