Highly Virulent and Multidrug-Resistant Escherichia coli Sequence Type 58 from a Sausage in Germany
- PMID: 35892394
- PMCID: PMC9331442
- DOI: 10.3390/antibiotics11081006
Highly Virulent and Multidrug-Resistant Escherichia coli Sequence Type 58 from a Sausage in Germany
Abstract
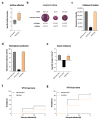
Studies have previously described the occurrence of multidrug-resistant (MDR) Escherichia coli in human and veterinary medical settings, livestock, and, to a lesser extent, in the environment and food. While they mostly analyzed foodborne E. coli regarding phenotypic and sometimes genotypic antibiotic resistance and basic phylogenetic classification, we have limited understanding of the in vitro and in vivo virulence characteristics and global phylogenetic contexts of these bacteria. Here, we investigated in-depth an E. coli strain (PBIO3502) isolated from a pork sausage in Germany in 2021. Whole-genome sequence analysis revealed sequence type (ST)58, which has an internationally emerging high-risk clonal lineage. In addition to its MDR phenotype that mostly matched the genotype, PBIO3502 demonstrated pronounced virulence features, including in vitro biofilm formation, siderophore secretion, serum resilience, and in vivo mortality in Galleria mellonella larvae. Along with the genomic analysis indicating close phylogenetic relatedness of our strain with publicly available, clinically relevant representatives of the same ST, these results suggest the zoonotic and pathogenic character of PBIO3502 with the potential to cause infection in humans and animals. Additionally, our study highlights the necessity of the One Health approach while integrating human, animal, and environmental health, as well as the role of meat products and food chains in the putative transmission of MDR pathogens.
Keywords: CTX-M-1; Enterobacterales; Escherichia coli; IncI1; One Health; antimicrobial resistance; food safety.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. - DOI - PubMed
-
- Ewers C., Bethe A., Semmler T., Guenther S., Wieler L.H. Extended-spectrum beta-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 2012;18:646–655. doi: 10.1111/j.1469-0691.2012.03850.x. - DOI - PubMed
LinkOut - more resources
Full Text Sources

