The Binding Landscape of Serum Antibodies: How Physical and Mathematical Concepts Can Advance Systems Immunology
- PMID: 35892703
- PMCID: PMC9326739
- DOI: 10.3390/antib11030043
The Binding Landscape of Serum Antibodies: How Physical and Mathematical Concepts Can Advance Systems Immunology
Abstract
Antibodies constitute a major component of serum on protein mass basis. We also know that the structural diversity of these antibodies exceeds that of all other proteins in the body and they react with an immense number of molecular targets. What we still cannot quantitatively describe is how antibody abundance is related to affinity, specificity, and cross reactivity. This ignorance has important practical consequences: we also do not have proper biochemical units for characterizing polyclonal serum antibody binding. The solution requires both a theoretical foundation, a physical model of the system, and technology for the experimental confirmation of theory. Here we argue that the quantitative characterization of interactions between serum antibodies and their targets requires systems-level physical chemistry approach and generates results that should help create maps of antibody binding landscape.
Keywords: antibody; chemical potential; differential equation; logistic function; system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

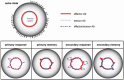

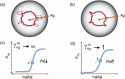
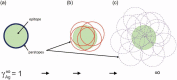
References
LinkOut - more resources
Full Text Sources

