MGRN1 as a Phenotypic Determinant of Human Melanoma Cells and a Potential Biomarker
- PMID: 35892921
- PMCID: PMC9331370
- DOI: 10.3390/life12081118
MGRN1 as a Phenotypic Determinant of Human Melanoma Cells and a Potential Biomarker
Abstract
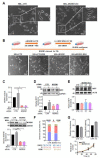
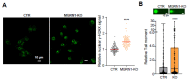
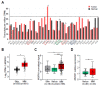
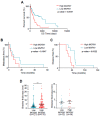
Mahogunin Ring Finger 1 (MGRN1), a ubiquitin ligase expressed in melanocytes, interacts with the α melanocyte-stimulating hormone receptor, a well-known melanoma susceptibility gene. Previous studies showed that MGRN1 modulates the phenotype of mouse melanocytes and melanoma cells, with effects on pigmentation, shape, and motility. Moreover, MGRN1 knockdown augmented the burden of DNA breaks in mouse cells, indicating that loss of MGRN1 promoted genomic instability. However, data concerning the roles of MGRN1 in human melanoma cells remain scarce. We analyzed MGRN1 knockdown in human melanoma cells. Transient MGRN1 depletion with siRNA or permanent knockdown in human melanoma cells by CRISPR/Cas9 caused an apparently MITF-independent switch to a more dendritic phenotype. Lack of MGRN1 also increased the fraction of human cells in the S phase of the cell cycle and the burden of DNA breaks but did not significantly impair proliferation. Moreover, in silico analysis of publicly available melanoma datasets and estimation of MGRN1 in a cohort of clinical specimens provided preliminary evidence that MGRN1 expression is higher in human melanomas than in normal skin or nevi and pointed to an inverse correlation of MGRN1 expression in human melanoma with patient survival, thus suggesting potential use of MGRN1 as a melanoma biomarker.
Keywords: DNA damage; Mahogunin Ring Finger 1 (MGRN1); biomarker; melanocytes; melanoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

