Global m6A RNA Methylation in SARS-CoV-2 Positive Nasopharyngeal Samples in a Mexican Population: A First Approximation Study
- PMID: 35893012
- PMCID: PMC9326742
- DOI: 10.3390/epigenomes6030016
Global m6A RNA Methylation in SARS-CoV-2 Positive Nasopharyngeal Samples in a Mexican Population: A First Approximation Study
Abstract
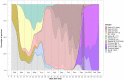
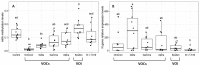
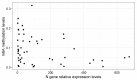
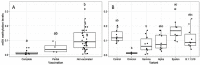
The Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2) is the causal agent of COVID-19 (Coronavirus Disease-19). Both mutation and/or recombination events in the SARS-CoV-2 genome have resulted in variants that differ in transmissibility and severity. Furthermore, RNA methylation of the N6 position of adenosine (m6A) is known to be altered in cells infected with SARS-CoV-2. However, it is not known whether this epitranscriptomic modification differs across individuals dependent on the presence of infection with distinct SARS-CoV-2 variants, the viral load, or the vaccination status. To address this issue, we selected RNAs (n = 60) from SARS-CoV-2 sequenced nasopharyngeal samples (n = 404) of 30- to 60-year-old outpatients or hospitalized individuals from the city of Mazatlán (Mexico) between February 2021 and March 2022. Control samples were non-infected individuals (n = 10). SARS-CoV-2 was determined with real-time PCR, viral variants were determined with sequencing, and global m6A levels were determined by using a competitive immunoassay method. We identified variants of concern (VOC; alpha, gamma, delta, omicron), the variant of interest (VOI; epsilon), and the lineage B.1.1.519. Global m6A methylation differed significantly across viral variants (p = 3.2 × 10-7). In particular, we found that m6A levels were significantly lower in the VOC delta- and omicron-positive individuals compared to non-infected individuals (p = 2.541236 × 10-2 and 1.134411 × 10-4, respectively). However, we uncovered no significant correlation between global m6A levels and viral nucleocapsid (N) gene expression or age. Furthermore, individuals with complete vaccination schemes showed significantly lower m6A levels than unvaccinated individuals (p = 2.6 × 10-4), and differences in methylation levels across variants in unvaccinated individuals were significant (p = 3.068 × 10-3). These preliminary results suggest that SARS-CoV-2 variants show differences in global m6A levels.
Keywords: SARS-CoV-2; m6A methylation; nasopharyngeal samples; viral variants.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. doi: 10.1126/science.abg3055. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

