Transcriptome and Metabonomic Analysis of Tamarix ramosissima Potassium (K+) Channels and Transporters in Response to NaCl Stress
- PMID: 35893048
- PMCID: PMC9394374
- DOI: 10.3390/genes13081313
Transcriptome and Metabonomic Analysis of Tamarix ramosissima Potassium (K+) Channels and Transporters in Response to NaCl Stress
Abstract
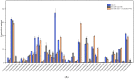
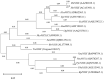
Potassium ion (K+) channels and transporters are key components of plant K+ absorption and transportation and play an important role in plant growth and development. This study revealed that K+ channels and transporters are involved in the salt tolerance molecular mechanism and metabolites of the halophyte representative plant Tamarix ramosissima (T. ramosissima) in response to NaCl stress, providing a theoretical basis for the mitigation of salt stress using halophytes. Through transcriptome sequencing and metabolite detection analysis of 0 h, 48 h and 168 h by applying exogenous K+ to the roots of T. ramosissima under NaCl stress, 15 high-quality Clean Data bases were obtained, Q20 reached more than 97%, Q30 reached more than 92%, and GC content reached 44.5%, which is in line with further bioinformatics analysis. Based on the Liquid chromatography−mass spectrometry (LC-MS) analysis, the roots of T. ramosissima were exposed to exogenous potassium for 48 h and 168 h under NaCl stress, and 1510 and 1124 metabolites were identified in positive and negative ion mode, respectively. Through orthogonal projections to latent structures discriminant analysis (OPLS-DA) model analysis, its metabolomic data have excellent predictability and stability. The results of this study showed that there were 37 differentially expressed genes (DEGs) annotated as Class 2 K+ channels (Shaker-like K+ channel and TPK channel) and Class 3 K+ transporters (HAK/KUP/KT, HKT and CPAs transporter families). Among them, 29 DEGs were annotated to the gene ontology (GO) database, and the most genes were involved in the GO Biological Process. In addition, the expression levels of Unigene0014342 in the HAK/KUP/KT transporter and Unigene0088276 and Unigene0103067 in the CPAs transporter both first decreased and then increased when treated with 200 mM NaCl for 48 h and 168 h. However, when treated with 200 mM NaCl + 10 mM KCl for 48 h and 168 h, a continuous upward trend was shown. Notably, the expression level of Unigene0016813 in CPAS transporter continued to increase when treated with 200 mM NaCl and 200 mM NaCl + 10 mM KCl for 48 h and 168 h. 3 DEGs, Unigene0088276, Unigene0016813 and Unigene0103067, were dominated by the positive regulation of their related metabolites, and this correlation was significant. The results showed that these DEGs increased the absorption of K+ and the ratio of K+/Na+ under NaCl stress at 48 h and 168 h after adding exogenous potassium and enhanced the salt tolerance of T. ramosissima. Notably, the expression level of Unigene0103067 in the CPAs transporter was consistently upregulated when 200 mM NaCl + 10 mM KCl was treated for 48 h and 168 h. The positive regulatory metabolites were always dominant, which better helped T. ramosissima resist salt stress. Unigene0103067 plays an important role in enhancing the salt tolerance of T. ramosissima and reducing the toxicity of NaCl in roots. Additionally, phylogenetic tree analysis showed that Unigene0103067 and Reaumuria trigyna had the closest genetic distance in the evolutionary relationship. Finally, 9 DEGs were randomly selected for quantitative real-time PCR (qRT-PCR) verification. Their expression trends were completely consistent with the transcriptome sequencing analysis results, proving that this study’s data are accurate and reliable. This study provides resources for revealing the molecular mechanism of NaCl stress tolerance in T. ramosissima and lays a theoretical foundation for cultivating new salt-tolerant varieties.
Keywords: NaCl stress; potassium channel; potassium ion transporter.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Leigh R.A., Jones R.G.W. A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol. 1984;97:1–13. doi: 10.1111/j.1469-8137.1984.tb04103.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

