Visual Clustering of Transcriptomic Data from Primary and Metastatic Tumors-Dependencies and Novel Pitfalls
- PMID: 35893071
- PMCID: PMC9394300
- DOI: 10.3390/genes13081335
Visual Clustering of Transcriptomic Data from Primary and Metastatic Tumors-Dependencies and Novel Pitfalls
Abstract
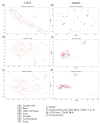
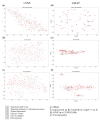
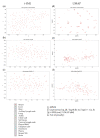
Personalized oncology is a rapidly evolving area and offers cancer patients therapy options that are more specific than ever. However, there is still a lack of understanding regarding transcriptomic similarities or differences of metastases and corresponding primary sites. Applying two unsupervised dimension reduction methods (t-Distributed Stochastic Neighbor Embedding (t-SNE) and Uniform Manifold Approximation and Projection (UMAP)) on three datasets of metastases (n =682 samples) with three different data transformations (unprocessed, log10 as well as log10 + 1 transformed values), we visualized potential underlying clusters. Additionally, we analyzed two datasets (n =616 samples) containing metastases and primary tumors of one entity, to point out potential familiarities. Using these methods, no tight link between the site of resection and cluster formation outcome could be demonstrated, or for datasets consisting of solely metastasis or mixed datasets. Instead, dimension reduction methods and data transformation significantly impacted visual clustering results. Our findings strongly suggest data transformation to be considered as another key element in the interpretation of visual clustering approaches along with initialization and different parameters. Furthermore, the results highlight the need for a more thorough examination of parameters used in the analysis of clusters.
Keywords: UMAP; cancer; metastasis; t-SNE; transcriptomic analysis; visual clustering.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Thomas R.M., Truty M.J., Nogueras-Gonzalez G.M., Fleming J.B., Vauthey J.-N., Pisters P.W.T., Lee J.E., Rice D.C., Hofstetter W.L., Wolff R.A., et al. Selective reoperation for locally recurrent or metastatic pancreatic ductal adenocarcinoma following primary pancreatic resection. J. Gastrointest. Surg. 2012;16:1696–1704. doi: 10.1007/s11605-012-1912-8. - DOI - PMC - PubMed
-
- Nishizaki T., DeVries S., Chew K., Goodson W.H., Ljung B.-M., Thor A., Waldman F.M. Genetic alterations in primary breast cancers and their metastases: Direct comparison using modified comparative genomic hybridization. Genes Chromosom. Cancer. 1997;19:267–272. doi: 10.1002/(SICI)1098-2264(199708)19:43.0.CO;2-V. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

