Systematic Analysis of Long Non-Coding RNA Genes in Nonalcoholic Fatty Liver Disease
- PMID: 35893239
- PMCID: PMC9332188
- DOI: 10.3390/ncrna8040056
Systematic Analysis of Long Non-Coding RNA Genes in Nonalcoholic Fatty Liver Disease
Abstract
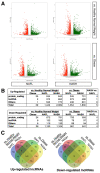
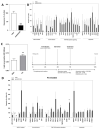
The largest solid organ in humans, the liver, performs a variety of functions to sustain life. When damaged, cells in the liver can regenerate themselves to maintain normal liver physiology. However, some damage is beyond repair, which necessitates liver transplantation. Increasing rates of obesity, Western diets (i.e., rich in processed carbohydrates and saturated fats), and cardiometabolic diseases are interlinked to liver diseases, including non-alcoholic fatty liver disease (NAFLD), which is a collective term to describe the excess accumulation of fat in the liver of people who drink little to no alcohol. Alarmingly, the prevalence of NAFLD extends to 25% of the world population, which calls for the urgent need to understand the disease mechanism of NAFLD. Here, we performed secondary analyses of published RNA sequencing (RNA-seq) data of NAFLD patients compared to healthy and obese individuals to identify long non-coding RNAs (lncRNAs) that may underly the disease mechanism of NAFLD. Similar to protein-coding genes, many lncRNAs are dysregulated in NAFLD patients compared to healthy and obese individuals, suggesting that understanding the functions of dysregulated lncRNAs may shed light on the pathology of NAFLD. To demonstrate the functional importance of lncRNAs in the liver, loss-of-function experiments were performed for one NAFLD-related lncRNA, LINC01639, which showed that it is involved in the regulation of genes related to apoptosis, TNF/TGF, cytokine signaling, and growth factors as well as genes upregulated in NAFLD. Since there is no lncRNA database focused on the liver, especially NAFLD, we built a web database, LiverDB, to further facilitate functional and mechanistic studies of hepatic lncRNAs.
Keywords: NAFLD; NASH; RNA-seq; gene expression; liver; lncRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

