Prevalence of pvmrp1 Polymorphisms and Its Contribution to Antimalarial Response
- PMID: 35893540
- PMCID: PMC9394237
- DOI: 10.3390/microorganisms10081482
Prevalence of pvmrp1 Polymorphisms and Its Contribution to Antimalarial Response
Abstract
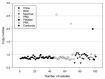
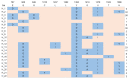
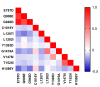
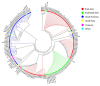
As more sporadic cases of chloroquine resistance occur (CQR) in Plasmodium vivax (P. vivax) malaria, molecular markers have become an important tool to monitor the introduction and spread of drug resistance. P. vivax multidrug resistance-associated protein 1 (PvMRP1), as one of the members of the ATP-binding cassette (ABC) transporters, may modulate this phenotype. In this study, we investigated the gene mutations and copy number variations (CNVs) in the pvmrp1 in 102 P. vivax isolates from China, the Republic of Korea (ROK), Myanmar, Papua New Guinea (PNG), Pakistan, the Democratic People’s Republic of Korea (PRK), and Cambodia. And we also obtained 72 available global pvmrp1 sequences deposited in the PlasmoDB database to investigate the genetic diversity, haplotype diversity, natural selection, and population structure of pvmrp1. In total, 29 single nucleotide polymorphisms reflected in 23 non-synonymous, five synonymous mutations and one gene deletion were identified, and CNVs were found in 2.9% of the isolates. Combined with the antimalarial drug susceptibility observed in the previous in vitro assays, except the prevalence of S354N between the two CQ sensitivity categories revealed a significant difference, no genetic mutations or CNVs associated with drug sensitivity were found. The genetic polymorphism analysis of 166 isolates worldwide found that the overall nucleotide diversity (π) of pvmrp1 was 0.0011, with 46 haplotypes identified (Hd = 0.9290). The ratio of non-synonymous to synonymous mutations (dn/ds = 0.5536) and the neutrality tests statistic Fu and Li’s D* test (Fu and Li’s D* = −3.9871, p < 0.02) suggests that pvmrp1 had evolved under a purifying selection. Due to geographical differences, genetic differentiation levels of pvmrp1 in different regions were different to some extent. Overall, this study provides a new idea for finding CQR molecular monitoring of P. vivax and provides more sequences of pvmrp1 in Asia for subsequent research. However, further validation is still needed through laboratory and epidemiological field studies of P. vivax samples from more regions.
Keywords: Plasmodium vivax; genetic diversity; natural selection; pvmrp1.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- WHO World Malaria Report 2021. [(accessed on 6 December 2021)]. Available online: https://www.who.int/publications/i/item/9789240040496.
-
- Battle K.E., Lucas T.C.D., Nguyen M., Howes R.E., Nandi A.K., Twohig K.A., Pfeffer D.A., Cameron E., Rao P.C., Casey D., et al. Mapping the global endemicity and clinical burden of Plasmodium vivax, 2000–2017: A spatial and temporal modelling study. Lancet. 2019;394:332–343. doi: 10.1016/S0140-6736(19)31096-7. - DOI - PMC - PubMed
-
- Xu S., Zeng W., Ngassa Mbenda H.G., Liu H., Chen X., Xiang Z., Li C., Zhang Y., Baird J.K., Yang Z., et al. Efficacy of directly-observed chloroquine-primaquine treatment for uncomplicated acute Plasmodium vivax malaria in northeast Myanmar: A prospective open-label efficacy trial. Travel Med. Infect. Dis. 2020;36:101499. doi: 10.1016/j.tmaid.2019.101499. - DOI - PMC - PubMed
Grants and funding
- BK20130114/the Natural Science Foundation of Jiangsu Province
- 2019-YY-065/the Six Talent Peaks Project in Jiangsu Province
- KYCX21-3284/the Postgraduate Research & Practice Innovation Program of Jiangsu Province
- NRF-2021R1A2C2008235/a National Research Foundation of Korea (NRF) grant
- NRF-R1A4A1031574/the Basic Science Research Program through the National Research Foundation of Korea (NRF)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

