Imaging Intron Evolution
- PMID: 35893579
- PMCID: PMC9326662
- DOI: 10.3390/mps5040053
Imaging Intron Evolution
Abstract
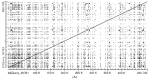
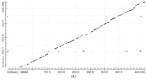
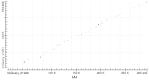
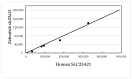
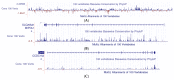
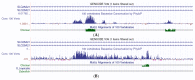
Intron evolution may be readily imaged through the combined use of the "dot plot" function of the NCBI BLAST, aligning two sequences at a time, and the Vertebrate "Multiz" alignment and conservation tool of the UCSC Genome Browser. With the NCBI BLAST, an ideal alignment of two highly conserved sequences generates a diagonal straight line in the plot from the lower left corner to the upper right corner. Gaps in this line correspond to non-conserved sections. In addition, the dot plot of the alignment of a sequence with the same sequence after the removal of the Transposable Elements (TEs) can be observed along the diagonal gaps that correspond to the sites of TE insertion. The UCSC Genome Browser can graph, along the entire sequence of a single gene, the level of overall conservation in vertebrates. This level can be compared with the conservation level of the gene in one or more selected vertebrate species. As an example, we show the graphic analysis of the intron conservation in two genes: the mitochondrial solute carrier 21 (SLC25A21) and the growth hormone receptor (GHR), whose coding sequences are conserved through vertebrates, while their introns show dramatic changes in nucleotide composition and even length. In the SLC25A21, a few short but significant nucleotide sequences are conserved in zebrafish, Xenopus and humans, and the rate of conservation steadily increases from chicken/human to mouse/human alignments. In the GHR, a less conserved gene, the earlier indication of intron conservation is a small signal in chicken/human alignment. The UCSC tool may simultaneously display the conservation level of a gene in different vertebrates, with reference to the level of overall conservation in Vertebrates. It is shown that, at least in SLC25A21, the sites of higher conservation are not always coincident in chicken and zebrafish nor are the sites of higher vertebrate conservation.
Keywords: BLAST dot plots; GHR; Multiz alignment and conservation tool; SLC25A21; intron evolution.
Conflict of interest statement
The authors state no conflict of interest.
Figures




References
-
- Matlin A.J., Moore M.J. Spliceosome assembly and composition. Adv. Exp. Med. Biol. 2007;623:14–35. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

