Immune Escape Associated with RBD Omicron Mutations and SARS-CoV-2 Evolution Dynamics
- PMID: 35893668
- PMCID: PMC9394476
- DOI: 10.3390/v14081603
Immune Escape Associated with RBD Omicron Mutations and SARS-CoV-2 Evolution Dynamics
Abstract
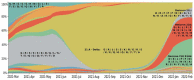
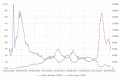
The evolution and the emergence of new mutations of viruses affect their transmissibility and/or pathogenicity features, depending on different evolutionary scenarios of virus adaptation to the host. A typical trade-off scenario of SARS-CoV-2 evolution has been proposed, which leads to the appearance of an Omicron strain with lowered lethality, yet enhanced transmissibility. This direction of evolution might be partly explained by virus adaptation to therapeutic agents and enhanced escape from vaccine-induced and natural immunity formed by other SARS-CoV-2 strains. Omicron's high mutation rate in the Spike protein, as well as its previously described high genome mutation rate (Kandeel et al., 2021), revealed a gap between it and other SARS-CoV-2 strains, indicating the absence of a transitional evolutionary form to the Omicron strain. Therefore, Omicron has emerged as a new serotype divergent from the evolutionary lineage of other SARS-CoV-2 strains. Omicron is a rapidly evolving variant of high concern, whose new subvariants continue to manifest. Its further understanding and the further monitoring of key mutations that provide virus immune escape and/or high affinity towards the receptor could be useful for vaccine and therapeutic development in order to control the evolutionary direction of the COVID-19 pandemic.
Keywords: COVID-19; Omicron; RBD mutations; SARS-CoV-2; bioinformatics; immune escape; vaccine development.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
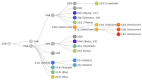
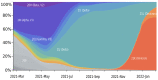
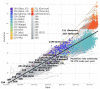
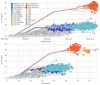
References
-
- WHO Coronavirus (COVID-19) Dashboard. [(accessed on 14 May 2022)]. Available online: https://covid19.who.int/
-
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

