Characterization of the cluster MabR prophages of Mycobacterium abscessus and Mycobacterium chelonae
- PMID: 35894699
- PMCID: PMC9434293
- DOI: 10.1093/g3journal/jkac188
Characterization of the cluster MabR prophages of Mycobacterium abscessus and Mycobacterium chelonae
Abstract
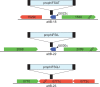
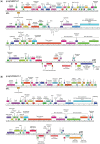
Mycobacterium abscessus is an emerging pathogen of concern in cystic fibrosis and immunocompromised patients and is considered one of the most drug-resistant mycobacteria. The majority of clinical Mycobacterium abscessus isolates carry 1 or more prophages that are hypothesized to contribute to virulence and bacterial fitness. The prophage McProf was identified in the genome of the Bergey strain of Mycobacterium chelonae and is distinct from previously described prophages of Mycobacterium abscessus. The McProf genome increases intrinsic antibiotic resistance of Mycobacterium chelonae and drives expression of the intrinsic antibiotic resistance gene, whiB7, when superinfected by a second phage. The prevalence of McProf-like genomes was determined in sequenced mycobacterial genomes. Related prophage genomes were identified in the genomes of 25 clinical isolates of Mycobacterium abscessus and assigned to the novel cluster, MabR. They share less than 10% gene content with previously described prophages; however, they share features typical of prophages, including polymorphic toxin-immunity systems.
Keywords: Mycobacterium; bacteriophage; genome; prophage.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
