Genetic architecture of dispersal and local adaptation drives accelerating range expansions
- PMID: 35895682
- PMCID: PMC9353510
- DOI: 10.1073/pnas.2121858119
Genetic architecture of dispersal and local adaptation drives accelerating range expansions
Abstract
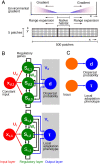
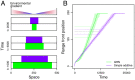
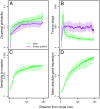
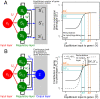
Contemporary evolution has the potential to significantly alter biotic responses to global change, including range expansion dynamics and biological invasions. Models predicting range dynamics often make highly simplifying assumptions about the genetic architecture underlying relevant traits. However, genetic architecture defines evolvability and higher-order evolutionary processes, which determine whether evolution will be able to keep up with environmental change or not. Therefore, we here study the impact of the genetic architecture of dispersal and local adaptation, two central traits of high relevance for range expansions, on the dynamics and predictability of invasion into an environmental gradient, such as temperature. In our theoretical model we assume that dispersal and local adaptation traits result from the products of two noninteracting gene-regulatory networks (GRNs). We compare our model to simpler quantitative genetics models and show that in the GRN model, range expansions are accelerating and less predictable. We further find that accelerating dynamics in the GRN model are primarily driven by an increase in the rate of local adaptation to novel habitats which results from greater sensitivity to mutation (decreased robustness) and increased gene expression. Our results highlight how processes at microscopic scales, here within genomes, can impact the predictions of large-scale, macroscopic phenomena, such as range expansions, by modulating the rate of evolution.
Keywords: biological invasion; environmental gradient; evolvability; gene-regulatory network; robustness.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Bullock J. M., et al., Human-mediated dispersal and the rewiring of spatial networks. Trends Ecol. Evol. 33, 958–970 (2018). - PubMed
-
- Chen I. C., Hill J. K., Ohlemüller R., Roy D. B., Thomas C. D., Rapid range shifts of species associated with high levels of climate warming. Science 333, 1024–1026 (2011). - PubMed
-
- Melbourne B. A., Hastings A., Highly variable spread rates in replicated biological invasions: Fundamental limits to predictability. Science 325, 1536–1539 (2009). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

