Butyrate acts through HDAC inhibition to enhance aryl hydrocarbon receptor activation by gut microbiota-derived ligands
- PMID: 35895845
- PMCID: PMC9336500
- DOI: 10.1080/19490976.2022.2105637
Butyrate acts through HDAC inhibition to enhance aryl hydrocarbon receptor activation by gut microbiota-derived ligands
Abstract
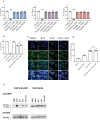
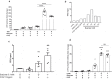
Aryl hydrocarbon receptor (AhR) is a critical player in the crosstalk between the gut microbiota and its host. However, factors regulating AhR within the gut, which is a complex metabolomic environment, are poorly understood. This study investigates the effect of a combination of metabolites on the activation mechanism of AhR. AhR activity was evaluated using both a luciferase reporter system and mRNA levels of AhR target genes on human cell lines and human colonic explants. AhR activation was studied by radioligand-binding assay, nuclear translocation of AhR by immuofluorescence and protein co-immunoprecipitation of AhR with ARNT. Indirect activation of AhR was evaluated using several tests and inhibitors. The promoter of the target gene CYP1A1 was studied both by chromatin immunoprecipitation and by using an histone deacetylase HDAC inhibitor (iHDAC). Short-chain fatty acids, and butyrate in particular, enhance AhR activity mediated by endogenous tryptophan metabolites without binding to the receptor. This effect was confirmed in human intestinal explants and did not rely on activation of receptors targeted by SCFAs, inhibition of AhR degradation or clearance of its ligands. Butyrate acted directly on AhR target gene promoter to reshape chromatin through iHDAC activity. Our findings revealed that butyrate is not an AhR ligand but acts as iHDAC leading to an increase recruitment of AhR to the target gene promoter in the presence of tryptophan-derived AhR agonists. These data contribute to a novel understanding of the complex regulation of AhR activation by gut microbiota-derived metabolites.
Keywords: AhR; CYP1A1; FICZ; HDAC; Microbiota; SCFAs; butyrate; metabolites; tryptophan.
Conflict of interest statement
HS report lecture fee, board membership, or consultancy from Carenity, AbbVie, Astellas, Danone, Ferring, Mayoly Spindler, MSD, Novartis, Roche, Tillots, Enterome, BiomX, Biose, Novartis,Takeda, Biocodex and is co-founder of Exeliom Biosciences.
JHL reports lecture fees from Ethicon, Takeda, Intuitive, B-Braun, invitation to a medical congress by Biomup, Intuitive and MD start. He is a consultant for Safeheal, Coloplast and FSK. He is a personal investor in 1 digital companies, medical device companies or biotech companies. Other authors declare no competing interests.
Figures


References
-
- Reigstad CS, Salmonson CE, Rainey JF, Szurszewski JH, Linden DR, Sonnenburg JL, Farrugia G, Kashyap PC.. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015;29(4):1395–1403. doi: 10.1096/fj.14-259598 - DOI - PMC - PubMed
-
- Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall H-U, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metabol. 2013;17(2):225–235. doi: 10.1016/j.cmet.2013.01.003 - DOI - PubMed
-
- Martin-Gallausiaux C, Larraufie P, Jarry A, Béguet-Crespel F, Marinelli L, Ledue F, Reimann F, Blottière HM, Lapaque N. Butyrate produced by commensal bacteria down-regulates indolamine 2,3-dioxygenase 1 (IDO-1) expression via a dual mechanism in human intestinal epithelial cells. Front Immunol. 2018;9:2838. doi: 10.3389/fimmu.2018.02838 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
