Differential neutralization and inhibition of SARS-CoV-2 variants by antibodies elicited by COVID-19 mRNA vaccines
- PMID: 35896523
- PMCID: PMC9328008
- DOI: 10.1038/s41467-022-31929-6
Differential neutralization and inhibition of SARS-CoV-2 variants by antibodies elicited by COVID-19 mRNA vaccines
Abstract
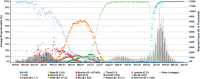
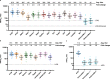
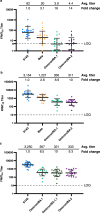
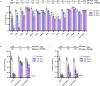
The evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in the emergence of new variant lineages that have exacerbated the COVID-19 pandemic. Some of those variants were designated as variants of concern/interest (VOC/VOI) by national or international authorities based on many factors including their potential impact on vaccine-mediated protection from disease. To ascertain and rank the risk of VOCs and VOIs, we analyze the ability of 14 variants (614G, Alpha, Beta, Gamma, Delta, Epsilon, Zeta, Eta, Theta, Iota, Kappa, Lambda, Mu, and Omicron) to escape from mRNA vaccine-induced antibodies. The variants show differential reductions in neutralization and replication by post-vaccination sera. Although the Omicron variant (BA.1, BA.1.1, and BA.2) shows the most escape from neutralization, sera collected after a third dose of vaccine (booster sera) retain moderate neutralizing activity against that variant. Therefore, vaccination remains an effective strategy during the COVID-19 pandemic.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
X.X. and P.-Y.S. have filed a patent on the reverse genetic system. X.X., and P.-Y.S. received compensation from Pfizer for COVID-19 vaccine development. Other authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

