Rare loss of function variants in the hepatokine gene INHBE protect from abdominal obesity
- PMID: 35896531
- PMCID: PMC9329324
- DOI: 10.1038/s41467-022-31757-8
Rare loss of function variants in the hepatokine gene INHBE protect from abdominal obesity
Abstract
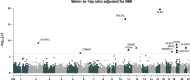
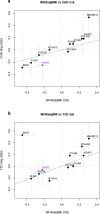
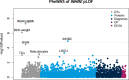
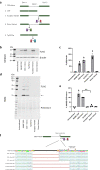
Identifying genetic variants associated with lower waist-to-hip ratio can reveal new therapeutic targets for abdominal obesity. We use exome sequences from 362,679 individuals to identify genes associated with waist-to-hip ratio adjusted for BMI (WHRadjBMI), a surrogate for abdominal fat that is causally linked to type 2 diabetes and coronary heart disease. Predicted loss of function (pLOF) variants in INHBE associate with lower WHRadjBMI and this association replicates in data from AMP-T2D-GENES. INHBE encodes a secreted protein, the hepatokine activin E. In vitro characterization of the most common INHBE pLOF variant in our study, indicates an in-frame deletion resulting in a 90% reduction in secreted protein levels. We detect associations with lower WHRadjBMI for variants in ACVR1C, encoding an activin receptor, further highlighting the involvement of activins in regulating fat distribution. These findings highlight activin E as a potential therapeutic target for abdominal obesity, a phenotype linked to cardiometabolic disease.
© 2022. The Author(s).
Conflict of interest statement
A.Deaton, A.Dubey, L.D.W., E.Y., S.T., L.N., R.A.H., C.W., M.E.P., A.M.H., K.F., A.K.V. and P.N. are employees and stockholders of Alnylam Pharmaceuticals. G.H. is an employee and stockholder of Alnylam Pharmaceuticals and a paid consultant to 54Gene. M.M.P. is a former employee and stockholder of Alnylam Pharmaceuticals and a current employee and stockholder of Beam Therapeutics. As of April 2022, P.D. is an employee and stockholder of Regeneron Pharmaceuticals. J.F. has no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

