SARS-CoV-2 nucleocapsid protein triggers hyperinflammation via protein-protein interaction-mediated intracellular Cl- accumulation in respiratory epithelium
- PMID: 35896532
- PMCID: PMC9328007
- DOI: 10.1038/s41392-022-01048-1
SARS-CoV-2 nucleocapsid protein triggers hyperinflammation via protein-protein interaction-mediated intracellular Cl- accumulation in respiratory epithelium
Abstract
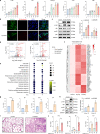
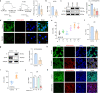
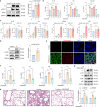
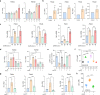
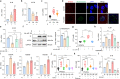
SARS-CoV-2, the culprit pathogen of COVID-19, elicits prominent immune responses and cytokine storms. Intracellular Cl- is a crucial regulator of host defense, whereas the role of Cl- signaling pathway in modulating pulmonary inflammation associated with SARS-CoV-2 infection remains unclear. By using human respiratory epithelial cell lines, primary cultured human airway epithelial cells, and murine models of viral structural protein stimulation and SARS-CoV-2 direct challenge, we demonstrated that SARS-CoV-2 nucleocapsid (N) protein could interact with Smad3, which downregulated cystic fibrosis transmembrane conductance regulator (CFTR) expression via microRNA-145. The intracellular Cl- concentration ([Cl-]i) was raised, resulting in phosphorylation of serum glucocorticoid regulated kinase 1 (SGK1) and robust inflammatory responses. Inhibition or knockout of SGK1 abrogated the N protein-elicited airway inflammation. Moreover, N protein promoted a sustained elevation of [Cl-]i by depleting intracellular cAMP via upregulation of phosphodiesterase 4 (PDE4). Rolipram, a selective PDE4 inhibitor, countered airway inflammation by reducing [Cl-]i. Our findings suggested that Cl- acted as the crucial pathological second messenger mediating the inflammatory responses after SARS-CoV-2 infection. Targeting the Cl- signaling pathway might be a novel therapeutic strategy for COVID-19.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

