Actin nano-architecture of phagocytic podosomes
- PMID: 35896550
- PMCID: PMC9329332
- DOI: 10.1038/s41467-022-32038-0
Actin nano-architecture of phagocytic podosomes
Abstract
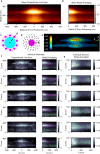
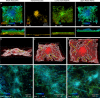
Podosomes are actin-enriched adhesion structures important for multiple cellular processes, including migration, bone remodeling, and phagocytosis. Here, we characterize the structure and organization of phagocytic podosomes using interferometric photoactivated localization microscopy, a super-resolution microscopy technique capable of 15-20 nm resolution, together with structured illumination microscopy and localization-based super-resolution microscopy. Phagocytic podosomes are observed during frustrated phagocytosis, a model in which cells attempt to engulf micropatterned IgG antibodies. For circular patterns, this results in regular arrays of podosomes with well-defined geometry. Using persistent homology, we develop a pipeline for semi-automatic identification and measurement of podosome features. These studies reveal an hourglass shape of the podosome actin core, a protruding knob at the bottom of the core, and two actin networks extending from the core. Additionally, the distributions of paxillin, talin, myosin II, α-actinin, cortactin, and microtubules relative to actin are characterized.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

