Integrated genomic sequencing in myeloid blast crisis chronic myeloid leukemia (MBC-CML), identified potentially important findings in the context of leukemogenesis model
- PMID: 35896598
- PMCID: PMC9329277
- DOI: 10.1038/s41598-022-17232-w
Integrated genomic sequencing in myeloid blast crisis chronic myeloid leukemia (MBC-CML), identified potentially important findings in the context of leukemogenesis model
Erratum in
-
Author Correction: Integrated genomic sequencing in myeloid blast crisis chronic myeloid leukemia (MBC-CML), identified potentially important findings in the context of leukemogenesis model.Sci Rep. 2023 Jan 13;13(1):736. doi: 10.1038/s41598-023-27872-1. Sci Rep. 2023. PMID: 36639723 Free PMC article. No abstract available.
Abstract
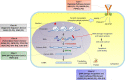
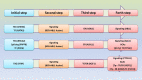
Chronic myeloid leukemia (CML) is a model of leukemogenesis in which the exact molecular mechanisms underlying blast crisis still remained unexplored. The current study identified multiple common and rare important findings in myeloid blast crisis CML (MBC-CML) using integrated genomic sequencing, covering all classes of genes implicated in the leukemogenesis model. Integrated genomic sequencing via Whole Exome Sequencing (WES), Chromosome-seq and RNA-sequencing were conducted on the peripheral blood samples of three CML patients in the myeloid blast crisis. An in-house filtering pipeline was applied to assess important variants in cancer-related genes. Standard variant interpretation guidelines were used for the interpretation of potentially important findings (PIFs) and potentially actionable findings (PAFs). Single nucleotide variation (SNV) and small InDel analysis by WES detected sixteen PIFs affecting all five known classes of leukemogenic genes in myeloid malignancies including signaling pathway components (ABL1, PIK3CB, PTPN11), transcription factors (GATA2, PHF6, IKZF1, WT1), epigenetic regulators (ASXL1), tumor suppressor and DNA repair genes (BRCA2, ATM, CHEK2) and components of spliceosome (PRPF8). These variants affect genes involved in leukemia stem cell proliferation, self-renewal, and differentiation. Both patients No.1 and No.2 had actionable known missense variants on ABL1 (p.Y272H, p.F359V) and frameshift variants on ASXL1 (p.A627Gfs*8, p.G646Wfs*12). The GATA2-L359S in patient No.1, PTPN11-G503V and IKZF1-R208Q variants in the patient No.3 were also PAFs. RNA-sequencing was used to confirm all of the identified variants. In the patient No. 3, chromosome sequencing revealed multiple pathogenic deletions in the short and long arms of chromosome 7, affecting at least three critical leukemogenic genes (IKZF1, EZH2, and CUX1). The large deletion discovered on the short arm of chromosome 17 in patient No. 2 resulted in the deletion of TP53 gene as well. Integrated genomic sequencing combined with RNA-sequencing can successfully discover and confirm a wide range of variants, from SNVs to CNVs. This strategy may be an effective method for identifying actionable findings and understanding the pathophysiological mechanisms underlying MBC-CML, as well as providing further insights into the genetic basis of MBC-CML and its management in the future.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

