Dairying, diseases and the evolution of lactase persistence in Europe
- PMID: 35896751
- PMCID: PMC7615474
- DOI: 10.1038/s41586-022-05010-7
Dairying, diseases and the evolution of lactase persistence in Europe
Erratum in
-
Author Correction: Dairying, diseases and the evolution of lactase persistence in Europe.Nature. 2022 Sep;609(7927):E9. doi: 10.1038/s41586-022-05227-6. Nature. 2022. PMID: 36042335 No abstract available.
Abstract
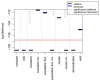
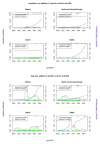
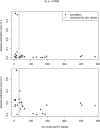
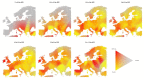
In European and many African, Middle Eastern and southern Asian populations, lactase persistence (LP) is the most strongly selected monogenic trait to have evolved over the past 10,000 years1. Although the selection of LP and the consumption of prehistoric milk must be linked, considerable uncertainty remains concerning their spatiotemporal configuration and specific interactions2,3. Here we provide detailed distributions of milk exploitation across Europe over the past 9,000 years using around 7,000 pottery fat residues from more than 550 archaeological sites. European milk use was widespread from the Neolithic period onwards but varied spatially and temporally in intensity. Notably, LP selection varying with levels of prehistoric milk exploitation is no better at explaining LP allele frequency trajectories than uniform selection since the Neolithic period. In the UK Biobank4,5 cohort of 500,000 contemporary Europeans, LP genotype was only weakly associated with milk consumption and did not show consistent associations with improved fitness or health indicators. This suggests that other reasons for the beneficial effects of LP should be considered for its rapid frequency increase. We propose that lactase non-persistent individuals consumed milk when it became available but, under conditions of famine and/or increased pathogen exposure, this was disadvantageous, driving LP selection in prehistoric Europe. Comparison of model likelihoods indicates that population fluctuations, settlement density and wild animal exploitation-proxies for these drivers-provide better explanations of LP selection than the extent of milk exploitation. These findings offer new perspectives on prehistoric milk exploitation and LP evolution.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors have no competing interests.
Figures















Comment in
-
How humans' ability to digest milk evolved from famine and disease.Nature. 2022 Aug;608(7922):251-252. doi: 10.1038/d41586-022-02067-2. Nature. 2022. PMID: 35896667 No abstract available.
-
The mystery of early milk consumption in Europe.Nature. 2022 Aug;608(7922):268-269. doi: 10.1038/d41586-022-02041-y. Nature. 2022. PMID: 35896670 No abstract available.
References
-
- Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006;312:1614. - PubMed
-
- Evershed RP, et al. Earliest date for milk use in the Near East and southeastern Europe linked to cattle herding. Nature. 2008;455:528–531. - PubMed
-
- Collins R. What makes UK Biobank special? Lancet. 2012;379:1173–1174. - PubMed
-
- Allen NE, Sudlow C, Peakman T, Collins R. UK Biobank Data: Come and Get It. Sci Transl Med. 2014;6:224ed224 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

