Molecular Mechanisms of Drug Resistance in Staphylococcus aureus
- PMID: 35897667
- PMCID: PMC9332259
- DOI: 10.3390/ijms23158088
Molecular Mechanisms of Drug Resistance in Staphylococcus aureus
Abstract
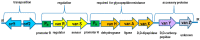
This paper discusses the mechanisms of S. aureus drug resistance including: (1) introduction. (2) resistance to beta-lactam antibiotics, with particular emphasis on the mec genes found in the Staphylococcaceae family, the structure and occurrence of SCCmec cassettes, as well as differences in the presence of some virulence genes and its expression in major epidemiological types and clones of HA-MRSA, CA-MRSA, and LA-MRSA strains. Other mechanisms of resistance to beta-lactam antibiotics will also be discussed, such as mutations in the gdpP gene, BORSA or MODSA phenotypes, as well as resistance to ceftobiprole and ceftaroline. (3) Resistance to glycopeptides (VRSA, VISA, hVISA strains, vancomycin tolerance). (4) Resistance to oxazolidinones (mutational and enzymatic resistance to linezolid). (5) Resistance to MLS-B (macrolides, lincosamides, ketolides, and streptogramin B). (6) Aminoglycosides and spectinomicin, including resistance genes, their regulation and localization (plasmids, transposons, class I integrons, SCCmec), and types and spectrum of enzymes that inactivate aminoglycosides. (7). Fluoroquinolones (8) Tetracyclines, including the mechanisms of active protection of the drug target site and active efflux of the drug from the bacterial cell. (9) Mupirocin. (10) Fusidic acid. (11) Daptomycin. (12) Resistance to other antibiotics and chemioterapeutics (e.g., streptogramins A, quinupristin/dalfopristin, chloramphenicol, rifampicin, fosfomycin, trimethoprim) (13) Molecular epidemiology of MRSA.
Keywords: HA/CA/LA-MRSA clones; SCCmec; Staphylococcus aureus; mechanisms of drug resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
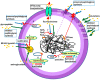
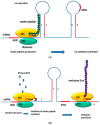
 , mechanisms of action—green arrows. Resistance to beta lactams: 1. Production of penicillin-binding protein PBP2A, 2. * mutations in PBP genes—rare (MODSA), 3. beta-lactamases production -usually narrow substrate spectrum. Glycopeptide resistance: 4. VanA operon (modification of the antibiotic binding site), Linezolid resistance: 5. adenylyl-N-methyltransferase Cfr-modification 23S rRNA of bacterial ribosome. Resistance to MLS-B (macrolides, lincosamides and streptogramins B): 5. Erm—erythromycin ribosome methylation. Aminoglycosides resistance: 6. antibiotics inactivation by tansferases. Fluoroinolones resistance: 7. mutations in gyrA and gyrB (topoisomerase II) and parC (grlA) and parE (topoisomerase IV) genes (modification of the antibiotic binding site), 8. removal from the bacterial cell by the efflux pump.
, mechanisms of action—green arrows. Resistance to beta lactams: 1. Production of penicillin-binding protein PBP2A, 2. * mutations in PBP genes—rare (MODSA), 3. beta-lactamases production -usually narrow substrate spectrum. Glycopeptide resistance: 4. VanA operon (modification of the antibiotic binding site), Linezolid resistance: 5. adenylyl-N-methyltransferase Cfr-modification 23S rRNA of bacterial ribosome. Resistance to MLS-B (macrolides, lincosamides and streptogramins B): 5. Erm—erythromycin ribosome methylation. Aminoglycosides resistance: 6. antibiotics inactivation by tansferases. Fluoroinolones resistance: 7. mutations in gyrA and gyrB (topoisomerase II) and parC (grlA) and parE (topoisomerase IV) genes (modification of the antibiotic binding site), 8. removal from the bacterial cell by the efflux pump.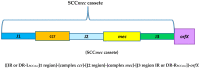
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

