The AP2/ERF GmERF113 Positively Regulates the Drought Response by Activating GmPR10-1 in Soybean
- PMID: 35897735
- PMCID: PMC9330420
- DOI: 10.3390/ijms23158159
The AP2/ERF GmERF113 Positively Regulates the Drought Response by Activating GmPR10-1 in Soybean
Abstract
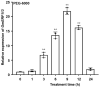
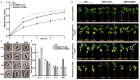
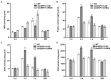
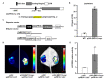
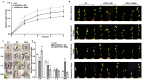
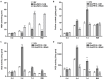
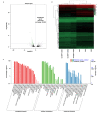
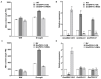
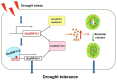
Ethylene response factors (ERFs) are involved in biotic and abiotic stress; however, the drought resistance mechanisms of many ERFs in soybeans have not been resolved. Previously, we proved that GmERF113 enhances resistance to the pathogen Phytophthora sojae in soybean. Here, we determined that GmERF113 is induced by 20% PEG-6000. Compared to the wild-type plants, soybean plants overexpressing GmERF113 (GmERF113-OE) displayed increased drought tolerance which was characterized by milder leaf wilting, less water loss from detached leaves, smaller stomatal aperture, lower Malondialdehyde (MDA) content, increased proline accumulation, and higher Superoxide dismutase (SOD) and Peroxidase (POD) activities under drought stress, whereas plants with GmERF113 silenced through RNA interference were the opposite. Chromatin immunoprecipitation and dual effector-reporter assays showed that GmERF113 binds to the GCC-box in the GmPR10-1 promoter, activating GmPR10-1 expression directly. Overexpressing GmPR10-1 improved drought resistance in the composite soybean plants with transgenic hairy roots. RNA-seq analysis revealed that GmERF113 downregulates abscisic acid 8'-hydroxylase 3 (GmABA8'-OH 3) and upregulates various drought-related genes. Overexpressing GmERF113 and GmPR10-1 increased the abscisic acid (ABA) content and reduced the expression of GmABA8'-OH3 in transgenic soybean plants and hairy roots, respectively. These results reveal that the GmERF113-GmPR10-1 pathway improves drought resistance and affects the ABA content in soybean, providing a theoretical basis for the molecular breeding of drought-tolerant soybean.
Keywords: ABA; GmERF113; GmPR10-1; drought tolerance; soybean.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
GmMKK4-activated GmMPK6 stimulates GmERF113 to trigger resistance to Phytophthora sojae in soybean.Plant J. 2022 Jul;111(2):473-495. doi: 10.1111/tpj.15809. Epub 2022 Jun 2. Plant J. 2022. PMID: 35562858
-
A Novel Soybean ERF Transcription Factor, GmERF113, Increases Resistance to Phytophthora sojae Infection in Soybean.Front Plant Sci. 2017 Mar 7;8:299. doi: 10.3389/fpls.2017.00299. eCollection 2017. Front Plant Sci. 2017. PMID: 28326092 Free PMC article.
-
Drought Tolerance Conferred in Soybean (Glycine max. L) by GmMYB84, a Novel R2R3-MYB Transcription Factor.Plant Cell Physiol. 2017 Oct 1;58(10):1764-1776. doi: 10.1093/pcp/pcx111. Plant Cell Physiol. 2017. PMID: 29016915
-
Proteomics and phosphoproteomics reveal the different drought-responsive mechanisms of priming with (Z)-3-hexenyl acetate in two tea cultivars.J Proteomics. 2023 Oct 30;289:105010. doi: 10.1016/j.jprot.2023.105010. Epub 2023 Oct 4. J Proteomics. 2023. PMID: 37797878 Review.
-
Transcriptional Stress Memory and Transgenerational Inheritance of Drought Tolerance in Plants.Int J Mol Sci. 2022 Oct 26;23(21):12918. doi: 10.3390/ijms232112918. Int J Mol Sci. 2022. PMID: 36361708 Free PMC article. Review.
Cited by
-
Physiological Characteristic Changes and Transcriptome Analysis of Maize (Zea mays L.) Roots under Drought Stress.Int J Genomics. 2024 Jan 17;2024:5681174. doi: 10.1155/2024/5681174. eCollection 2024. Int J Genomics. 2024. PMID: 38269194 Free PMC article.
-
Dissection of quantitative trait nucleotides and candidate genes associated with agronomic and yield-related traits under drought stress in rapeseed varieties: integration of genome-wide association study and transcriptomic analysis.Front Plant Sci. 2024 Mar 19;15:1342359. doi: 10.3389/fpls.2024.1342359. eCollection 2024. Front Plant Sci. 2024. PMID: 38567131 Free PMC article.
-
Identification of nuclear factor YA6 genes in sorghum and characterization of their involvement in drought tolerance.Front Plant Sci. 2025 Mar 19;16:1524066. doi: 10.3389/fpls.2025.1524066. eCollection 2025. Front Plant Sci. 2025. PMID: 40177019 Free PMC article.
-
Transcriptome Analysis Reveals Differentially Expressed Genes Involved in Cadmium and Arsenic Accumulation in Tea Plant (Camellia sinensis).Plants (Basel). 2023 Mar 5;12(5):1182. doi: 10.3390/plants12051182. Plants (Basel). 2023. PMID: 36904042 Free PMC article.
-
A wild-allele GsPP2C-51-a1 enhances tolerance to drought stress in soybean and Arabidopsis.Theor Appl Genet. 2025 Feb 24;138(3):51. doi: 10.1007/s00122-025-04835-w. Theor Appl Genet. 2025. PMID: 39994030
References
-
- Zhou Y.Y., Zhang Y., Wang X.W., Han X., An Y., Lin S.W., Shen C., Wen J., Liu C., Yin W., et al. The root-specific NF-Y family transcription factor, PdNF-YB21, positively regulates root growth and drought resistance by ABA-mediated IAA transport in Populus. New Phytol. 2020;227:407–426. doi: 10.1111/nph.16524. - DOI - PubMed
-
- Nahar K., Hasanuzzaman M., Fujita M. Osmolytes and Plants Acclimation to Changing Environment: Emerging Omics Technologies. Springer; New Delhi, India: 2016. Roles of osmolytes in plant adaptation to drought and salinity; pp. 37–68. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

