Characterization of SARS-CoV-2 Escape Mutants to a Pair of Neutralizing Antibodies Targeting the RBD and the NTD
- PMID: 35897753
- PMCID: PMC9332373
- DOI: 10.3390/ijms23158177
Characterization of SARS-CoV-2 Escape Mutants to a Pair of Neutralizing Antibodies Targeting the RBD and the NTD
Abstract
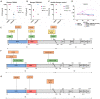
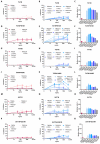
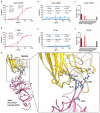
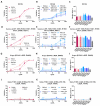
Mutations in the spike protein of SARS-CoV-2 can lead to evasion from neutralizing antibodies and affect the efficacy of passive and active immunization strategies. Immunization of mice harboring an entire set of human immunoglobulin variable region gene segments allowed to identify nine neutralizing monoclonal antibodies, which either belong to a cluster of clonally related RBD or NTD binding antibodies. To better understand the genetic barrier to emergence of SARS-CoV-2 variants resistant to these antibodies, escape mutants were selected in cell culture to one antibody from each cluster and a combination of the two antibodies. Three independently derived escape mutants to the RBD antibody harbored mutations in the RBD at the position T478 or S477. These mutations impaired the binding of the RBD antibodies to the spike protein and conferred resistance in a pseudotype neutralization assay. Although the binding of the NTD cluster antibodies were not affected by the RBD mutations, the RBD mutations also reduced the neutralization efficacy of the NTD cluster antibodies. The mutations found in the escape variants to the NTD antibody conferred resistance to the NTD, but not to the RBD cluster antibodies. A variant resistant to both antibodies was more difficult to select and only emerged after longer passages and higher inoculation volumes. VOC carrying the same mutations as the ones identified in the escape variants were also resistant to neutralization. This study further underlines the rapid emergence of escape mutants to neutralizing monoclonal antibodies in cell culture and indicates the need for thorough investigation of escape mutations to select the most potent combination of monoclonal antibodies for clinical use.
Keywords: NTD; RBD; SARS-CoV-2; SARS-CoV-2 escape mutations; neutralizing monoclonal antibodies; spike protein; variants of concern.
Conflict of interest statement
K.Ü. has filed a patent application on TRES antibodies and holds shares of COVER Antibodies GmbH, a company that has licensed IP rights on TRES antibodies. The other authors declare no commercial or financial conflict of interest.
Figures

Similar articles
-
A Combination of Receptor-Binding Domain and N-Terminal Domain Neutralizing Antibodies Limits the Generation of SARS-CoV-2 Spike Neutralization-Escape Mutants.mBio. 2021 Oct 26;12(5):e0247321. doi: 10.1128/mBio.02473-21. Epub 2021 Oct 5. mBio. 2021. PMID: 34607456 Free PMC article.
-
Emergence of Multiple SARS-CoV-2 Antibody Escape Variants in an Immunocompromised Host Undergoing Convalescent Plasma Treatment.mSphere. 2021 Aug 25;6(4):e0048021. doi: 10.1128/mSphere.00480-21. Epub 2021 Aug 25. mSphere. 2021. PMID: 34431691 Free PMC article.
-
Neutralization potency of monoclonal antibodies recognizing dominant and subdominant epitopes on SARS-CoV-2 Spike is impacted by the B.1.1.7 variant.Immunity. 2021 Jun 8;54(6):1276-1289.e6. doi: 10.1016/j.immuni.2021.03.023. Epub 2021 Apr 1. Immunity. 2021. PMID: 33836142 Free PMC article.
-
SARS-CoV-2 Variants of Concern and Variants of Interest Receptor Binding Domain Mutations and Virus Infectivity.Front Immunol. 2022 Jan 27;13:825256. doi: 10.3389/fimmu.2022.825256. eCollection 2022. Front Immunol. 2022. PMID: 35154144 Free PMC article. Review.
-
Is SARS-CoV-2 Spike Evolution Being Retargeted at the N-Terminal Domain?Discov Med. 2025 May;37(196):801-807. doi: 10.24976/Discov.Med.202537196.70. Discov Med. 2025. PMID: 40415355 Review.
Cited by
-
Influence of AAV vector tropism on long-term expression and Fc-γ receptor binding of an antibody targeting SARS-CoV-2.Commun Biol. 2024 Jul 16;7(1):865. doi: 10.1038/s42003-024-06529-3. Commun Biol. 2024. PMID: 39009807 Free PMC article.
-
Status and Developing Strategies for Neutralizing Monoclonal Antibody Therapy in the Omicron Era of COVID-19.Viruses. 2023 May 31;15(6):1297. doi: 10.3390/v15061297. Viruses. 2023. PMID: 37376597 Free PMC article. Review.
-
Propagation of SARS-CoV-2 in a Closed Cell Culture Device: Potential GMP Compatible Production Platform for Live-Attenuated Vaccine Candidates under BSL-3 Conditions?Viruses. 2023 Jan 30;15(2):397. doi: 10.3390/v15020397. Viruses. 2023. PMID: 36851610 Free PMC article.
-
Genetic barrier to resistance: a critical parameter for efficacy of neutralizing monoclonal antibodies against SARS-CoV-2 in a nonhuman primate model.J Virol. 2024 Jul 23;98(7):e0062824. doi: 10.1128/jvi.00628-24. Epub 2024 Jun 20. J Virol. 2024. PMID: 38899895 Free PMC article.
-
SARS-CoV-2 Spike Protein Is Capable of Inducing Cell-Cell Fusions Independent from Its Receptor ACE2 and This Activity Can Be Impaired by Furin Inhibitors or a Subset of Monoclonal Antibodies.Viruses. 2023 Jul 4;15(7):1500. doi: 10.3390/v15071500. Viruses. 2023. PMID: 37515187 Free PMC article.
References
-
- Cov-Lineages Lineage List. 2021 [20.11.2021 29.11.2021] [(accessed on 21 December 2021)]. Available online: https://cov-lineages.org/lineage_list.html.
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

