DNA Damage Response Regulation by Histone Ubiquitination
- PMID: 35897775
- PMCID: PMC9332593
- DOI: 10.3390/ijms23158187
DNA Damage Response Regulation by Histone Ubiquitination
Abstract
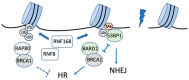
Cells are constantly exposed to numerous genotoxic stresses that induce DNA damage. DNA double-strand breaks (DSBs) are among the most serious damages and should be systematically repaired to preserve genomic integrity. The efficiency of repair is closely associated with chromatin structure, which is regulated by posttranslational modifications of histones, including ubiquitination. Recent evidence shows crosstalk between histone ubiquitination and DNA damage responses, suggesting an integrated model for the systematic regulation of DNA repair. There are two major pathways for DSB repair, viz., nonhomologous end joining and homologous recombination, and the choice of the pathway is partially controlled by posttranslational modifications of histones, including ubiquitination. Histone ubiquitination changes chromatin structure in the vicinity of DSBs and serves as a platform to select and recruit repair proteins; the removal of these modifications by deubiquitinating enzymes suppresses the recruitment of repair proteins and promotes the convergence of repair reactions. This article provides a comprehensive overview of the DNA damage response regulated by histone ubiquitination in response to DSBs.
Keywords: DNA repair; chromatin; histone modifications; ubiquitination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

