A Genome-Wide Association Study Coupled With a Transcriptomic Analysis Reveals the Genetic Loci and Candidate Genes Governing the Flowering Time in Alfalfa (Medicago sativa L.)
- PMID: 35898229
- PMCID: PMC9310038
- DOI: 10.3389/fpls.2022.913947
A Genome-Wide Association Study Coupled With a Transcriptomic Analysis Reveals the Genetic Loci and Candidate Genes Governing the Flowering Time in Alfalfa (Medicago sativa L.)
Abstract
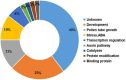
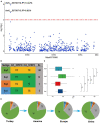
The transition to flowering at the right time is very important for adapting to local conditions and maximizing alfalfa yield. However, the understanding of the genetic basis of the alfalfa flowering time remains limited. There are few reliable genes or markers for selection, which hinders progress in genetic research and molecular breeding of this trait in alfalfa. We sequenced 220 alfalfa cultivars and conducted a genome-wide association study (GWAS) involving 875,023 single-nucleotide polymorphisms (SNPs). The phenotypic analysis showed that the breeding status and geographical origin strongly influenced the alfalfa flowering time. Our GWAS revealed 63 loci significantly related to the flowering time. Ninety-five candidate genes were detected at these SNP loci within 40 kb (20 kb up- and downstream). Thirty-six percent of the candidate genes are involved in development and pollen tube growth, indicating that these genes are key genetic mechanisms of alfalfa growth and development. The transcriptomic analysis showed that 1,924, 2,405, and 3,779 differentially expressed genes (DEGs) were upregulated across the three growth stages, while 1,651, 2,613, and 4,730 DEGs were downregulated across the stages. Combining the results of our GWAS and transcriptome analysis, in total, 38 candidate genes (7 differentially expressed during the bud stage, 13 differentially expressed during the initial flowering stage, and 18 differentially expressed during the full flowering stage) were identified. Two SNPs located in the upstream region of the Msa0888690 gene (which is involved in isop renoids) were significantly related to flowering. The two significant SNPs within the upstream region of Msa0888690 existed as four different haplotypes in this panel. The genes identified in this study represent a series of candidate targets for further research investigating the alfalfa flowering time and could be used for alfalfa molecular breeding.
Keywords: GWAS; SNP; alfalfa; flowering time; haplotypes.
Copyright © 2022 He, Zhang, Jiang, Long, Wang, Chen, Li, Gao, Yang, Wang, Kang, Chen and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Genome-Wide Association Analysis Coupled With Transcriptome Analysis Reveals Candidate Genes Related to Salt Stress in Alfalfa (Medicago sativa L.).Front Plant Sci. 2022 Feb 3;12:826584. doi: 10.3389/fpls.2021.826584. eCollection 2021. Front Plant Sci. 2022. PMID: 35185967 Free PMC article.
-
QTL mapping of flowering time and biomass yield in tetraploid alfalfa (Medicago sativa L.).BMC Plant Biol. 2019 Aug 16;19(1):359. doi: 10.1186/s12870-019-1946-0. BMC Plant Biol. 2019. PMID: 31419945 Free PMC article.
-
Transcriptome and GWAS Analyses Reveal Candidate Gene for Root Traits of Alfalfa during Germination under Salt Stress.Int J Mol Sci. 2023 Mar 27;24(7):6271. doi: 10.3390/ijms24076271. Int J Mol Sci. 2023. PMID: 37047244 Free PMC article.
-
Transcriptomic analysis of differentially expressed genes in leaves and roots of two alfalfa (Medicago sativa L.) cultivars with different salt tolerance.BMC Plant Biol. 2021 Oct 5;21(1):446. doi: 10.1186/s12870-021-03201-4. BMC Plant Biol. 2021. PMID: 34610811 Free PMC article.
-
A genome-wide association study reveals novel loci and candidate genes associated with plant height variation in Medicago sativa.BMC Plant Biol. 2024 Jun 13;24(1):544. doi: 10.1186/s12870-024-05151-z. BMC Plant Biol. 2024. PMID: 38872112 Free PMC article.
Cited by
-
Advances in basic biology of alfalfa (Medicago sativa L.): a comprehensive overview.Hortic Res. 2025 Mar 10;12(7):uhaf081. doi: 10.1093/hr/uhaf081. eCollection 2025 Jul. Hortic Res. 2025. PMID: 40343348 Free PMC article.
-
The Genetic Dissection of Nitrogen Use-Related Traits in Flax (Linum usitatissimum L.) at the Seedling Stage through the Integration of Multi-Locus GWAS, RNA-seq and Genomic Selection.Int J Mol Sci. 2023 Dec 18;24(24):17624. doi: 10.3390/ijms242417624. Int J Mol Sci. 2023. PMID: 38139451 Free PMC article.
-
MODMS: a multi-omics database for facilitating biological studies on alfalfa (Medicago sativa L.).Hortic Res. 2023 Nov 27;11(1):uhad245. doi: 10.1093/hr/uhad245. eCollection 2024 Jan. Hortic Res. 2023. PMID: 38239810 Free PMC article.
-
Analyzing Medicago spp. seed morphology using GWAS and machine learning.Sci Rep. 2024 Jul 30;14(1):17588. doi: 10.1038/s41598-024-67790-4. Sci Rep. 2024. PMID: 39080407 Free PMC article.
References
-
- Annicchiarico P., Barrett B., Brummer E. C., Julier B., Marshall A. H. (2015). Achievements and challenges in improving temperate perennial forage legumes. Crit. Rev. Plant Sci. 34 327–380.
-
- Arzani H., Zohdi M., Fish E., Amiri G. H. Z., Nikkhah A., Wester D. (2004). Phenological effects on forage quality of five grass species. J. Range Manage. 57 624–629.
LinkOut - more resources
Full Text Sources

