Characterization of Immune-Related Molecular Subtypes and a Prognostic Signature Correlating With the Response to Immunotherapy in Patients With Gastric Cancer
- PMID: 35898512
- PMCID: PMC9309259
- DOI: 10.3389/fimmu.2022.939836
Characterization of Immune-Related Molecular Subtypes and a Prognostic Signature Correlating With the Response to Immunotherapy in Patients With Gastric Cancer
Abstract
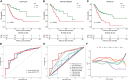
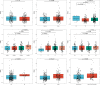
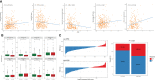
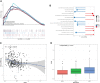
Gastric cancer (GC) is a disease characterized by high molecular and phenotypic heterogeneity and represents a leading cause of cancer-related death worldwide. The tumor immune microenvironment (TIME) affects the response to immunotherapy and the prognosis of patients with GC. Explorations of the TIME in GC and characterization of molecular subtypes might enhance personalized treatment and facilitate clinical decision-making. In this study, two molecular subtypes were defined through unsupervised consensus clustering based on immune-related dysregulated genes. Then, patients with different molecular subtypes of GC were shown to have distinct differences in sensitivity to immune checkpoint blockers (ICBs). The immune-related prognostic signature was established utilizing least absolute shrinkage and selection operator (LASSO)-Cox regression analysis. Three independent external cohorts and the IMvigor210 cohort were introduced to validate the robustness of IPRS. scRNA-seq data of GC samples were used to decipher the underlying mechanisms of how IPRS contributes to the TIME. GC biospecimens were collected for RT-qPCR to further validate our findings. In summary, we characterized the abnormal TIME of GC and constructed a reliable immune-related prognostic signature correlating with the response to immunotherapy. This study may provide new strategies for developing individualized treatments for patients with GC.
Keywords: gastric cancer; gene signature; immunotherapy; molecular subtypes; prognosis.
Copyright © 2022 Wang, Yang, Wang, Hu, Zhang, Guo, Chen, Jiang and Cui.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

