Immunoprotective Effects of Two Histone H2A Variants in the Grass Carp Against Flavobacterium columnare Infection
- PMID: 35898515
- PMCID: PMC9310644
- DOI: 10.3389/fimmu.2022.939464
Immunoprotective Effects of Two Histone H2A Variants in the Grass Carp Against Flavobacterium columnare Infection
Abstract
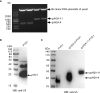
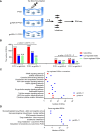
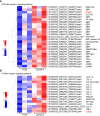
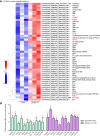
In teleost fish, the nucleotide polymorphisms of histone H2A significantly affect the resistance or susceptibility of zebrafish to Edwardsiella piscicida infection. Whether histone H2A variants can enhance the resistance of grass carp to Flavobacterium columnare infection remains unclear. Here, the effects of 7 previously obtained variants (gcH2A-1~gcH2A-7) and 5 novel histone H2A variants (gcH2A-11, gcH2A-13~gcH2A-16) in response to F. columnare infection were investigated. It was found that these histone H2A variants could be divided into type I and II. Among them, 5 histone H2A variants had no any effects on the F. columnare infection, however 7 histone H2A variants had antibacterial activity against F. columnare infection. The gcH2A-4 and gcH2A-11, whose antibacterial activity was the strongest in type I and II histone H2A variants respectively, were picked out for yeast expression. Transcriptome data for the samples from the intestines of grass carp immunized with the engineered Saccharomyces cerevisiae expressing PYD1, gcH2A-4 or gcH2A-11 revealed that 5 and 12 immune-related signaling pathways were significantly enriched by gcH2A-4 or gcH2A-11, respectively. For the engineered S. cerevisiae expressing gcH2A-4, NOD-like receptor and Toll-like receptor signaling pathways were enriched for up-regulated DEGs. Besides NOD-like receptor and Toll-like receptor signaling pathways, the engineered S. cerevisiae expressing gcH2A-11 also activated Cytosolic DNA-sensing pathway, RIG-I-like receptor signaling pathway and C-type lectin receptor signaling pathway. Furthermore, grass carp were immunized with the engineered S. cerevisiae expressing PYD1, gcH2A-4 or gcH2A-11 for 1 month and challenged with F. columnare. These grass carp immunized with gcH2A-4 or gcH2A-11 showed lower mortality and fewer numbers of F. columnare than did the control group. All these results suggest that gcH2A-4 and gcH2A-11 play important roles in evoking the innate immune responses and enhancing disease resistance of grass carp against F. columnare infection.
Keywords: Flavobacterium columnare; Saccharomyces cerevisiae; grass carp; histone H2A variants; immunoprotective effect.
Copyright © 2022 Yang, Zheng, Fang, Wu, Zhang and Chang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

