Cargo and Functional Profile of Saliva-Derived Exosomes Reveal Biomarkers Specific for Head and Neck Cancer
- PMID: 35899209
- PMCID: PMC9309685
- DOI: 10.3389/fmed.2022.904295
Cargo and Functional Profile of Saliva-Derived Exosomes Reveal Biomarkers Specific for Head and Neck Cancer
Abstract
Background: Exosomes contribute to immunosuppression in head and neck squamous cell carcinoma (HNSCC), a tumor entity which lacks specific tumor biomarkers. Plasma-derived exosomes from HNSCC patients correlate with clinical parameters and have potential as liquid biopsy. Here, we investigate the cargo and functional profile of saliva-derived exosomes from HNSCC patients and their potential as non-invasive biomarkers for disease detection and immunomodulation.
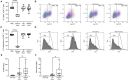
Methods: Exosomes were isolated from saliva of HNSCC patients (n = 21) and healthy donors (HD, n = 12) by differential ultracentrifugation. Surface values of immune checkpoints and tumor associated antigens on saliva-derived exosomes were analyzed by bead-based flow cytometry using CD63 capture. Upon co-incubation with saliva-derived exosomes, activity and proliferation of T cells were assessed by flow cytometry (CD69 expression, CFSE assay). Adenosine levels were measured by mass spectrometry after incubation of saliva-derived exosomes with exogenous ATP. miRNA profiling of saliva-derived exosomes was performed using the nCounter® SPRINT system.
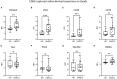
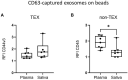
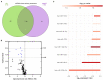
Results: Saliva-derived, CD63-captured exosomes from HNSCC patients carried high amounts of CD44v3, PDL1 and CD39. Compared to plasma, saliva was rich in tumor-derived, CD44v3+ exosomes and poor in hematopoietic cell-derived, CD45+ exosomes. CD8+ T cell activity was attenuated by saliva-derived exosomes from HNSCC patients, while proliferation of CD4+ T cells was not affected. Further, saliva-derived exosomes produced high levels of immunosuppressive adenosine. 62 HD- and 31 HNSCC-exclusive miRNAs were identified. Samples were grouped in "Healthy" and "Cancer" based on their saliva-derived exosomal miRNA profile, which was further found to be involved in RAS/MAPK, NF-κB complex, Smad2/3, and IFN-α signaling.
Conclusions: Saliva-derived exosomes from HNSCC patients were enriched in tumor-derived exosomes whose cargo and functional profile reflected an immunosuppressive TME. Surface values of CD44v3, PDL1 and CD39 on CD63-captured exosomes, adenosine production and the miRNA cargo of saliva-derived exosomes emerged as discriminators of disease and emphasized their potential as liquid biomarkers specific for HNSCC.
Keywords: exosomes; head and neck squamous cell carcinoma (HNSCC); liquid biopsy; miRNA; saliva.
Copyright © 2022 Hofmann, Medyany, Ezić, Lotfi, Niesler, Röth, Engelhardt, Laban, Schuler, Hoffmann, Brunner, Jackson and Theodoraki.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
[Advances in salivary exosomal miRNAs in head and neck squamous carcinoma].Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2024 Mar;38(3):261-266. doi: 10.13201/j.issn.2096-7993.2024.03.016. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2024. PMID: 38433699 Free PMC article. Review. Chinese.
-
Comparison of plasma- and saliva-derived exosomal miRNA profiles reveals diagnostic potential in head and neck cancer.Front Cell Dev Biol. 2022 Aug 22;10:971596. doi: 10.3389/fcell.2022.971596. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36072342 Free PMC article.
-
CD44v3 protein-carrying tumor-derived exosomes in HNSCC patients' plasma as potential noninvasive biomarkers of disease activity.Oncoimmunology. 2020 Apr 7;9(1):1747732. doi: 10.1080/2162402X.2020.1747732. eCollection 2020. Oncoimmunology. 2020. PMID: 32313730 Free PMC article.
-
Separation of plasma-derived exosomes into CD3(+) and CD3(-) fractions allows for association of immune cell and tumour cell markers with disease activity in HNSCC patients.Clin Exp Immunol. 2018 Jun;192(3):271-283. doi: 10.1111/cei.13113. Epub 2018 Mar 12. Clin Exp Immunol. 2018. PMID: 29431869 Free PMC article.
-
The Emerging Role of Exosomes in Diagnosis, Prognosis, and Therapy in Head and Neck Cancer.Int J Mol Sci. 2020 Jun 6;21(11):4072. doi: 10.3390/ijms21114072. Int J Mol Sci. 2020. PMID: 32517240 Free PMC article. Review.
Cited by
-
Comparative analyses of salivary exosomal miRNAs for patients with or without lung cancer.Front Genet. 2023 Nov 3;14:1249678. doi: 10.3389/fgene.2023.1249678. eCollection 2023. Front Genet. 2023. PMID: 38028609 Free PMC article.
-
Evaluation of Thermal Liquid Biopsy Analysis of Saliva and Blood Plasma Specimens as a Novel Diagnostic Modality in Head and Neck Cancer.Cancers (Basel). 2024 Dec 18;16(24):4220. doi: 10.3390/cancers16244220. Cancers (Basel). 2024. PMID: 39766119 Free PMC article.
-
[Advances in salivary exosomal miRNAs in head and neck squamous carcinoma].Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2024 Mar;38(3):261-266. doi: 10.13201/j.issn.2096-7993.2024.03.016. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2024. PMID: 38433699 Free PMC article. Review. Chinese.
-
Leveraging Saliva for Insights into Head and Neck Cancer.Int J Mol Sci. 2024 Dec 17;25(24):13514. doi: 10.3390/ijms252413514. Int J Mol Sci. 2024. PMID: 39769275 Free PMC article. Review.
-
Advances in Research on Isothermal Signal Amplification Mediated MicroRNA Detection of Clinical Samples: Application to Disease Diagnosis.Biosensors (Basel). 2025 Jun 18;15(6):395. doi: 10.3390/bios15060395. Biosensors (Basel). 2025. PMID: 40558477 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

