Genome-wide analysis of hybridization in wild boar populations reveals adaptive introgression from domestic pig
- PMID: 35899256
- PMCID: PMC9309462
- DOI: 10.1111/eva.13432
Genome-wide analysis of hybridization in wild boar populations reveals adaptive introgression from domestic pig
Abstract
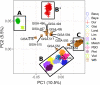
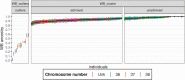
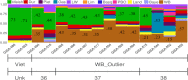
The admixture of domestic pig into French wild boar populations has been monitored since the 1980s thanks to the existence of a cytogenetic difference between the two sub-species. The number of chromosomes is 2n = 36 in wild boar and 2n = 38 in pig, respectively. This difference makes it possible to assign the "hybrid" status to wild boar individuals controlled with 37 or 38 chromosomes. However, it does not make it possible to determine the timing of the hybridization(s), nor to guarantee the absence of domestic admixture in an animal with 2n = 36 chromosomes. In order to analyze hybridization in greater detail and to avoid the inherent limitations of the cytogenetic approach, 362 wild boars (WB) recently collected in different French geographical areas and in different environments (farms, free ranging in protected or unprotected areas, animals with 2n = 36, 37 or 38 chromosomes) were genotyped on a 70K SNP chip. Principal component analyses allowed the identification of 13 "outliers" (3.6%), for which the proportion of the genome of "domestic" origin was greater than 40% (Admixture analyses). These animals were probably recent hybrids, having Asian domestic pig ancestry for most of them. For the remaining 349 animals studied, the proportion of the genome of "wild" origin varied between 83% and 100% (median: 94%). This proportion varied significantly depending on how the wild boar populations were managed. Local ancestry analyses revealed adaptive introgression from domestic pig, suggesting a critical role of genetic admixture in improving the fitness and population growth of WB. Overall, our results show that the methods used to monitor the domestic genetic contributions to wild boar populations should evolve in order to limit the level of admixture between the two gene pools.
Keywords: Sus scrofa; Wildlife management; admixture; feral pig; genetic monitoring; genotyping; population genetics.
© 2022 The Authors. Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non‐financial interest in the subject matter or materials discussed in this manuscript.
Figures

References
-
- Albrycht, M. , Merta, D. , Bobek, J. , & Ulejczyk, S. (2016). The demographic pattern of wild boars (Sus scrofa) inhabiting fragmented forest in North‐Eastern Poland. Baltic Forestry, 22(2), 251–258.
-
- Aravena, P. , & Skewes, O. (2007). European wild boar purebred and Sus scrofa intercrosses. Discrimination proposals. A review. Agro Ciencia, 23, 133–147.
-
- Barrios‐Garcia, M. N. , & Ballari, S. A. (2012). Impact of wild boar (Sus scrofa) in its introduced and native range: A review. Biological Invasions, 14(11), 2283–2300. 10.1007/s10530-012-0229-6 - DOI
LinkOut - more resources
Full Text Sources

