Analysis of rare driving events in pediatric acute myeloid leukemia
- PMID: 35899387
- PMCID: PMC9827169
- DOI: 10.3324/haematol.2021.280250
Analysis of rare driving events in pediatric acute myeloid leukemia
Abstract
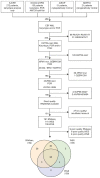
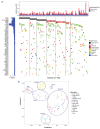
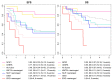
Elucidating genetic aberrations in pediatric acute myeloid leukemia (AML) provides insight in biology and may impact on risk-group stratification and clinical outcome. This study aimed to detect such aberrations in a selected series of samples without known (cyto)genetic aberration using molecular profiling. A cohort of 161 patients was selected from various study groups: DCOG, BFM, SJCRH, NOPHO and AEIOP. Samples were analyzed using RNA sequencing (n=152), whole exome (n=135) and/or whole genome sequencing (n=100). In 70 of 156 patients (45%), of whom RNA sequencing or whole genome sequencing was available, rearrangements were detected, 22 of which were novel; five involving ERG rearrangements and four NPM1 rearrangements. ERG rearrangements showed self-renewal capacity in vitro, and a distinct gene expression pattern. Gene set enrichment analysis of this cluster showed upregulation of gene sets derived from Ewing sarcoma, which was confirmed comparing gene expression profiles of AML and Ewing sarcoma. Furthermore, NPM1-rearranged cases showed cytoplasmic NPM1 localization and revealed HOXA/B gene overexpression, as described for NPM1 mutated cases. Single-gene mutations as identified in adult AML were rare. Patients had a median of 24 coding mutations (range, 7-159). Novel recurrent mutations were detected in UBTF (n=10), a regulator of RNA transcription. In 75% of patients an aberration with a prognostic impact could be detected. Therefore, we suggest these techniques need to become standard of care in diagnostics.
Figures


References
-
- Rubnitz JE, Gibson B, Smith FO. Acute myeloid leukemia. Hematol Oncol Clin North Am. 2010;24(1):35-63. - PubMed
-
- Rubnitz JE. Current management of childhood acute myeloid leukemia. Paediatr Drugs. 2017;19(1):1-10. - PubMed
-
- Creutzig U, van den Heuvel-Eibrink MM, Gibson B, et al. . Diagnosis and management of acute myeloid leukemia in children and adolescents: recommendations from an international expert panel. Blood. 2012;120(16):3187-3205. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

