Convergent biological pathways underlying the Kallmann syndrome-linked genes Hs6st1 and Fgfr1
- PMID: 35899427
- PMCID: PMC9759331
- DOI: 10.1093/hmg/ddac172
Convergent biological pathways underlying the Kallmann syndrome-linked genes Hs6st1 and Fgfr1
Abstract
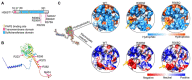
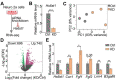
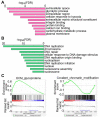
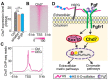
Kallmann syndrome (KS) is a congenital disorder characterized by idiopathic hypogonadotropic hypogonadism and olfactory dysfunction. KS is linked to variants in >34 genes, which are scattered across the human genome and show disparate biological functions. Although the genetic basis of KS is well studied, the mechanisms by which disruptions of these diverse genes cause the same outcome of KS are not fully understood. Here we show that disruptions of KS-linked genes affect the same biological processes, indicating convergent molecular mechanisms underlying KS. We carried out machine learning-based predictions and found that KS-linked mutations in heparan sulfate 6-O-sulfotransferase 1 (HS6ST1) are likely loss-of-function mutations. We next disrupted Hs6st1 and another KS-linked gene, fibroblast growth factor receptor 1 (Fgfr1), in mouse neuronal cells and measured transcriptome changes using RNA sequencing. We found that disruptions of Hs6st1 and Fgfr1 altered genes in the same biological processes, including the upregulation of genes in extracellular pathways and the downregulation of genes in chromatin pathways. Moreover, we performed genomics and bioinformatics analyses and found that Hs6st1 and Fgfr1 regulate gene transcription likely via the transcription factor Sox9/Sox10 and the chromatin regulator Chd7, which are also associated with KS. Together, our results demonstrate how different KS-linked genes work coordinately in a convergent signaling pathway to regulate the same biological processes, thus providing new insights into KS.
© The Author(s) 2022. Published by Oxford University Press.
Figures



Similar articles
-
Mutations in fibroblast growth factor receptor 1 cause Kallmann syndrome with a wide spectrum of reproductive phenotypes.Mol Cell Endocrinol. 2006 Jul 25;254-255:60-9. doi: 10.1016/j.mce.2006.04.021. Epub 2006 Jun 9. Mol Cell Endocrinol. 2006. PMID: 16764984
-
Prioritizing genetic testing in patients with Kallmann syndrome using clinical phenotypes.J Clin Endocrinol Metab. 2013 May;98(5):E943-53. doi: 10.1210/jc.2012-4116. Epub 2013 Mar 26. J Clin Endocrinol Metab. 2013. PMID: 23533228 Free PMC article.
-
Expanding the mutational spectrum of monogenic hypogonadotropic hypogonadism: novel mutations in ANOS1 and FGFR1 genes.Reprod Biol Endocrinol. 2020 Jan 29;18(1):8. doi: 10.1186/s12958-020-0568-6. Reprod Biol Endocrinol. 2020. PMID: 31996231 Free PMC article.
-
Diversity in fibroblast growth factor receptor 1 regulation: learning from the investigation of Kallmann syndrome.J Neuroendocrinol. 2008 Feb;20(2):141-63. doi: 10.1111/j.1365-2826.2007.01627.x. Epub 2007 Nov 22. J Neuroendocrinol. 2008. PMID: 18034870 Review.
-
Novel insights in FGFR1 regulation: lessons from Kallmann syndrome.Trends Endocrinol Metab. 2010 Jun;21(6):385-93. doi: 10.1016/j.tem.2010.01.004. Epub 2010 Feb 1. Trends Endocrinol Metab. 2010. PMID: 20117945 Review.
Cited by
-
Variety of genetic defects in GnRH and hypothalamic-pituitary signaling and development in normosmic patients with IHH.Front Endocrinol (Lausanne). 2024 Jul 1;15:1396805. doi: 10.3389/fendo.2024.1396805. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39010903 Free PMC article.
-
Vagal sensory neuron-derived FGF3 controls insulin secretion.Dev Cell. 2025 Jan 6;60(1):51-61.e4. doi: 10.1016/j.devcel.2024.09.016. Epub 2024 Oct 15. Dev Cell. 2025. PMID: 39413782
-
Knockout of the intellectual disability-linked gene Hs6st2 in mice decreases heparan sulfate 6-O-sulfation, impairs dendritic spines of hippocampal neurons, and affects memory.Glycobiology. 2024 Mar 26;34(2):cwad095. doi: 10.1093/glycob/cwad095. Glycobiology. 2024. PMID: 38015989 Free PMC article.
-
Spontaneous Improvement of Hypogonadotropic Hypogonadism in a Patient with PCSK1 and HS6ST1 Mutations: A Case Report.Life (Basel). 2025 Jul 21;15(7):1151. doi: 10.3390/life15071151. Life (Basel). 2025. PMID: 40724652 Free PMC article.
References
-
- Mitchell, A.L., Dwyer, A., Pitteloud, N. and Quinton, R. (2011) Genetic basis and variable phenotypic expression of Kallmann syndrome: towards a unifying theory. Trends Endocrinol. Metab., 22, 249–258. - PubMed
-
- Boehm, U., Bouloux, P.M., Dattani, M.T., de Roux, N., Dode, C., Dunkel, L., Dwyer, A.A., Giacobini, P., Hardelin, J.P., Juul, A. et al. (2015) Expert consensus document: European Consensus Statement on congenital hypogonadotropic hypogonadism--pathogenesis, diagnosis and treatment. Nat. Rev. Endocrinol., 11, 547–564. - PubMed
-
- Schwanzel-Fukuda, M., Bick, D. and Pfaff, D.W. (1989) Luteinizing hormone-releasing hormone (LHRH)-expressing cells do not migrate normally in an inherited hypogonadal (Kallmann) syndrome. Brain Res. Mol. Brain Res., 6, 311–326. - PubMed
-
- Kaluzna, M., Budny, B., Rabijewski, M., Kaluzny, J., Dubiel, A., Trofimiuk-Muldner, M., Wrotkowska, E., Hubalewska-Dydejczyk, A., Ruchala, M. and Ziemnicka, K. (2021) Defects in GnRH neuron migration/development and hypothalamic-pituitary signaling impact clinical variability of Kallmann syndrome. Genes (Basel), 12, 868. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

