Reduced Cathepsin L expression and secretion into the extracellular milieu contribute to lung fibrosis in systemic sclerosis
- PMID: 35900152
- PMCID: PMC10167927
- DOI: 10.1093/rheumatology/keac411
Reduced Cathepsin L expression and secretion into the extracellular milieu contribute to lung fibrosis in systemic sclerosis
Abstract
Objectives: Lung fibrosis is the leading cause of death in SSc, with no cure currently available. Antifibrotic Endostatin (ES) production does not reach therapeutic levels in SSc patients, suggesting a deficit in its release from Collagen XVIII by the main cleavage enzyme, Cathepsin L (CTSL). Thus, elucidating a potential deficit in CTSL expression and activity unravels an underlying molecular cause for SSc-driven lung fibrosis.
Methods: Fibrosis was induced experimentally using TGF-β in vitro, in primary human lung fibroblasts (pLFs), and ex vivo, in human lung tissues. ES and CTSL expression was quantified using ELISA, RT-qPCR, immunoblotting or immunofluorescence. Recombinant NC1-FLAG peptide was used to assess CTSL cleavage activity. CTSL expression was also compared between SSc vs normal (NL)-derived pLFs and lung tissues.
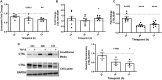
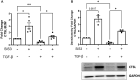
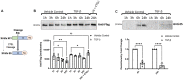
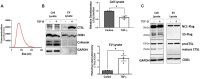
Results: ES levels were significantly reduced in media conditioned by TGF-β-induced pLFs. TGF-β-stimulated pLFs significantly reduced expression and secretion of CTSL into the extracellular matrix (ECM). CTSL was also sequestered in its inactive form into extracellular vesicles, further reducing its availability in the ECM. Media conditioned by TGF-β-induced pLFs showed reduced cleavage of NC1-Flag and reduced release of the antifibrotic ES fragment. SSc-derived pLFs and lung tissues expressed significantly lower levels of CTSL compared with NL.
Conclusions: Our findings identify CTSL as a protein protective against lung fibrosis via its activation of antifibrotic ES, and whose expression in SSc pLFs and lung tissues is suppressed. Identifying strategies to boost CTSL endogenous levels in SSc patients could serve as a viable therapeutic strategy.
Keywords: Cathepsin L; SSc; TGF-β; endostatin; extracellular vesicles; fibroblasts; fibrosis; lung; scleroderma.
© The Author(s) 2022. Published by Oxford University Press on behalf of the British Society for Rheumatology. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures


Comment in
-
Extracellular vesicle pathogenic cues in systemic sclerosis: comment on the article by Mouawad et al.Arthritis Rheumatol. 2024 Jan;76(1):148-149. doi: 10.1002/art.42684. Epub 2023 Oct 4. Arthritis Rheumatol. 2024. PMID: 37651269 No abstract available.
References
-
- Ho YY, Lagares D, Tager AM, Kapoor M.. Fibrosis–a lethal component of systemic sclerosis. Nat Rev Rheumatol 2014;10:390–402. - PubMed
-
- Elhai M, Meune C, Boubaya M. et al. ; EUSTAR group. Mapping and predicting mortality from systemic sclerosis. Ann Rheum Dis 2017;76:1897–905. - PubMed
-
- Khanna D, Lin CJF, Furst DE. et al. ; focuSSced investigators. Tocilizumab in systemic sclerosis: a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Respir Med 2020;8:963–74. - PubMed
-
- Distler O, Highland KB, Gahlemann M. et al. ; SENSCIS Trial Investigators. Nintedanib for systemic sclerosis-associated interstitial lung disease. N Engl J Med 2019;380:2518–28. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

