Small-molecule eRF3a degraders rescue CFTR nonsense mutations by promoting premature termination codon readthrough
- PMID: 35900863
- PMCID: PMC9479597
- DOI: 10.1172/JCI154571
Small-molecule eRF3a degraders rescue CFTR nonsense mutations by promoting premature termination codon readthrough
Abstract
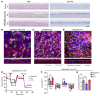
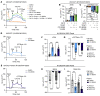
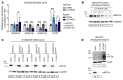
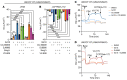
The vast majority of people with cystic fibrosis (CF) are now eligible for CF transmembrane regulator (CFTR) modulator therapy. The remaining individuals with CF harbor premature termination codons (PTCs) or rare CFTR variants with limited treatment options. Although the clinical modulator response can be reliably predicted using primary airway epithelial cells, primary cells carrying rare CFTR variants are scarce. To overcome this obstacle, cell lines can be created by overexpression of mouse Bmi-1 and human TERT (hTERT). Using this approach, we developed 2 non-CF and 6 CF airway epithelial cell lines, 3 of which were homozygous for the W1282X PTC variant. The Bmi-1/hTERT cell lines recapitulated primary cell morphology and ion transport function. The 2 F508del-CFTR cell lines responded robustly to CFTR modulators, which was mirrored in the parent primary cells and in the cell donors' clinical response. Cereblon E3 ligase modulators targeting eukaryotic release factor 3a (eRF3a) rescued W1282X-CFTR function to approximately 20% of WT levels and, when paired with G418, rescued G542X-CFTR function to approximately 50% of WT levels. Intriguingly, eRF3a degraders also diminished epithelial sodium channel (ENaC) function. These studies demonstrate that Bmi-1/hTERT cell lines faithfully mirrored primary cell responses to CFTR modulators and illustrate a therapeutic approach to rescue CFTR nonsense mutations.
Keywords: Genetic diseases; Ion channels; Pulmonology; Therapeutics; Translation.
Figures