ClinPharmSeq: A targeted sequencing panel for clinical pharmacogenetics implementation
- PMID: 35901010
- PMCID: PMC9333201
- DOI: 10.1371/journal.pone.0272129
ClinPharmSeq: A targeted sequencing panel for clinical pharmacogenetics implementation
Abstract
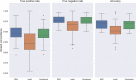
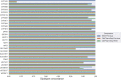
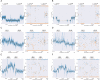
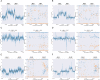
The accurate identification of genetic variants contributing to therapeutic drug response or adverse effects is the first step in implementation of precision drug therapy. Targeted sequencing has recently become a common methodology for large-scale studies of genetic variation thanks to its favorable balance between low cost, high throughput, and deep coverage. Here, we present ClinPharmSeq, a targeted sequencing panel of 59 genes with associations to pharmacogenetic (PGx) phenotypes, as a platform to explore the relationship between drug response and genetic variation, both common and rare. For validation, we sequenced DNA from 64 ethnically diverse Coriell samples with ClinPharmSeq to call star alleles (haplotype patterns) in 27 genes using the bioinformatics tool PyPGx. These reference samples were extensively characterized by multiple laboratories using PGx testing assays and, more recently, whole genome sequencing. We found that ClinPharmSeq can consistently generate deep-coverage data (mean = 274x) with high uniformity (30x or above = 94.8%). Our genotype analysis identified a total of 185 unique star alleles from sequencing data, and showed that diplotype calls from ClinPharmSeq are highly concordant with that from previous publications (97.6%) and whole genome sequencing (97.9%). Notably, all 19 star alleles with complex structural variation including gene deletions, duplications, and hybrids were recalled with 100% accuracy. Altogether, these results demonstrate that the ClinPharmSeq platform offers a feasible path for broad implementation of PGx testing and optimization of individual drug treatments.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Sanderson S, Emery J, Higgins J. CYP2C9 gene variants, drug dose, and bleeding risk in warfarin-treated patients: a HuGEnet systematic review and meta-analysis. Genet Med. 2005. Feb;7(2):97–104. doi: 10.1097/01.gim.0000153664.65759.cf - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

