Building the Plant SynBio Toolbox through Combinatorial Analysis of DNA Regulatory Elements
- PMID: 35901078
- PMCID: PMC9396662
- DOI: 10.1021/acssynbio.2c00147
Building the Plant SynBio Toolbox through Combinatorial Analysis of DNA Regulatory Elements
Abstract
While the installation of complex genetic circuits in microorganisms is relatively routine, the synthetic biology toolbox is severely limited in plants. Of particular concern is the absence of combinatorial analysis of regulatory elements, the long design-build-test cycles associated with transgenic plant analysis, and a lack of naming standardization for cloning parts. Here, we use previously described plant regulatory elements to design, build, and test 91 transgene cassettes for relative expression strength. Constructs were transiently transfected into Nicotiana benthamiana leaves and expression of a fluorescent reporter was measured from plant canopies, leaves, and protoplasts isolated from transfected plants. As anticipated, a dynamic level of expression was achieved from the library, ranging from near undetectable for the weakest cassette to a ∼200-fold increase for the strongest. Analysis of expression levels in plant canopies, individual leaves, and protoplasts were correlated, indicating that any of the methods could be used to evaluate regulatory elements in plants. Through this effort, a well-curated 37-member part library of plant regulatory elements was characterized, providing the necessary data to standardize construct design for precision metabolic engineering in plants.
Keywords: flow cytometry; fluorometry; genetic regulatory elements; single-cell analysis; synthetic biology; transgene expression.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

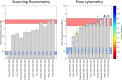





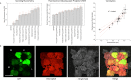
References
-
- Occhialini A.; Lin M. T.; Andralojc P. J.; Hanson M. R.; Parry M. A. Transgenic tobacco plants with improved cyanobacterial Rubisco expression but no extra assembly factors grow at near wild-type rates if provided with elevated CO2. Plant J. 2016, 85, 148–160. 10.1111/tpj.13098. - DOI - PMC - PubMed
-
- Fitch M. M. M.; Manshardt R. M.; Gonsalves D.; Slightom J. L.; Sanford J. C. Virus resistant papaya plants derived from tissues bombarded with the coat protein gene of papaya ringspot virus. Nat. Biotechnol. 1992, 10, 1466–1472. 10.1038/nbt1192-1466. - DOI
-
- Castiglioni P.; Warner D.; Bensen R. J.; Anstrom D. C.; Harrison J.; Stoecker M.; Abad M.; Kumar G.; Salvador S.; D’Ordine R.; Navarro S.; Back S.; Fernandes M.; Targolli J.; Dasgupta S.; Bonin C.; Luethy M. H.; Heard J. E. Bacterial RNA chaperones confer abiotic stress tolerance in plants and improved grain yield in maize under water-limited conditions. Plant Physiol. 2008, 147, 446–455. 10.1104/pp.108.118828. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

