The dynamic network of IS30 transposition pathways
- PMID: 35901099
- PMCID: PMC9333248
- DOI: 10.1371/journal.pone.0271414
The dynamic network of IS30 transposition pathways
Abstract
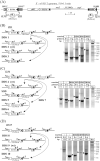
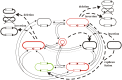
The E. coli element IS30 has adopted the copy-out-paste-in transposition mechanism that is prevalent in a number of IS-families. As an initial step, IS30 forms free circular transposition intermediates like IS minicircles or tandem IS-dimers by joining the inverted repeats of a single element or two, sometimes distantly positioned IS copies, respectively. Then, the active IR-IR junction of these intermediates reacts with the target DNA, which generates insertions, deletions, inversions or cointegrates. The element shows dual target specificity as it can insert into hot spot sequences or next to its inverted repeats. In this study the pathways of rearrangements of transposition-derived cointegrate-like structures were examined. The results showed that the probability of further rearrangements in these structures depends on whether the IS elements are flanked by hot spot sequences or take part in an IR-IR junction. The variability of the deriving products increases with the number of simultaneously available IRs and IR-IR joints in the cointegrates or the chromosome. Under certain conditions, the parental structures whose transposition formed the cointegrates are restored and persist among the rearranged products. Based on these findings, a novel dynamic model has been proposed for IS30, which possibly fits to other elements that have adopted the same transposition mechanism. The model integrates the known transposition pathways and the downstream rearrangements occurring after the formation of different cointegrate-like structures into a complex network. Important feature of this network is the presence of "feedback loops" and reversible transposition rearrangements that can explain how IS30 generates variability and preserves the original genetic constitution in the bacterial population, which contributes to the adaptability and evolution of host bacteria.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Formation and transposition of the covalently closed IS30 circle: the relation between tandem dimers and monomeric circles.Mol Microbiol. 1999 Oct;34(1):37-52. doi: 10.1046/j.1365-2958.1999.01567.x. Mol Microbiol. 1999. PMID: 10540284
-
Formation of the tandem repeat (IS30)2 and its role in IS30-mediated transpositional DNA rearrangements.Mol Gen Genet. 1993 May;239(1-2):177-87. doi: 10.1007/BF00281616. Mol Gen Genet. 1993. PMID: 8389976
-
Gene conversion in transposition of Escherichia coli element IS30.J Mol Biol. 2003 Dec 12;334(5):967-78. doi: 10.1016/j.jmb.2003.10.011. J Mol Biol. 2003. PMID: 14643660
-
Novel rearrangements of IS30 carrying plasmids leading to the reactivation of gene expression.Mol Gen Genet. 1987 May;207(2-3):413-20. doi: 10.1007/BF00331609. Mol Gen Genet. 1987. PMID: 3039299
-
Importance of illegitimate recombination and transposition in IS30-associated excision events.Plasmid. 1999 Nov;42(3):192-209. doi: 10.1006/plas.1999.1425. Plasmid. 1999. PMID: 10545262
Cited by
-
Differential Transcriptomic Profile of Piscirickettsia salmonis LF-89 and EM-90 During an In Vivo Spatial Separation Co-Culture in Atlantic Salmon.Microorganisms. 2024 Dec 2;12(12):2480. doi: 10.3390/microorganisms12122480. Microorganisms. 2024. PMID: 39770683 Free PMC article.
-
Genomic Characterization of Extended-Spectrum β-Lactamase (ESBL) Producing E. coli Harboring bla OXA-1-catB3-arr-3 Genes Isolated From Dairy Farm Environment in China.Transbound Emerg Dis. 2024 Oct 11;2024:3526395. doi: 10.1155/2024/3526395. eCollection 2024. Transbound Emerg Dis. 2024. PMID: 40303104 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

