Complete chloroplast genomes and phylogeny in three Euterpe palms (E. edulis, E. oleracea and E. precatoria) from different Brazilian biomes
- PMID: 35901127
- PMCID: PMC9333295
- DOI: 10.1371/journal.pone.0266304
Complete chloroplast genomes and phylogeny in three Euterpe palms (E. edulis, E. oleracea and E. precatoria) from different Brazilian biomes
Abstract
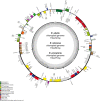
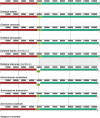
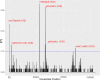
The Brazilian palm fruits and hearts-of-palm of Euterpe edulis, E. oleracea and E. precatoria are an important source for agro-industrial production, due to overexploitation, conservation strategies are required to maintain genetic diversity. Chloroplast genomes have conserved sequences, which are useful to explore evolutionary questions. Besides the plastid DNA, genome skimming allows the identification of other genomic resources, such as single nucleotide polymorphisms (SNPs), providing information about the genetic diversity of species. We sequenced the chloroplast genome and identified gene content in the three Euterpe species. We performed comparative analyses, described the polymorphisms among the chloroplast genome sequences (repeats, indels and SNPs) and performed a phylogenomic inference based on 55 palm species chloroplast genomes. Finally, using the remaining data from genome skimming, the nuclear and mitochondrial reads, we identified SNPs and estimated the genetic diversity among these Euterpe species. The Euterpe chloroplast genomes varied from 159,232 to 159,275 bp and presented a conserved quadripartite structure with high synteny with other palms. In a pairwise comparison, we found a greater number of insertions/deletions (indels = 93 and 103) and SNPs (284 and 254) between E. edulis/E. oleracea and E. edulis/E. precatoria when compared to E. oleracea/E. precatoria (58 indels and 114 SNPs). Also, the phylogeny indicated a closer relationship between E. oleracea/E. precatoria. The nuclear and mitochondrial genome analyses identified 1,077 SNPs and high divergence among species (FST = 0.77), especially between E. edulis and E. precatoria (FST = 0.86). These results showed that, despite the few structural differences among the chloroplast genomes of these Euterpe palms, a differentiation between E. edulis and the other Euterpe species can be identified by point mutations. This study not only brings new knowledge about the evolution of Euterpe chloroplast genomes, but also these new resources open the way for future phylogenomic inferences and comparative analyses within Arecaceae.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Baker WJ, Couvreur TLP. Global biogeography and diversification of palms sheds light on the evolution of tropical lineages. I. Historical biogeography. J Biogeogr. 2013;40(2): 274–285. doi: 10.1111/j.1365-2699.2012.02795.x - DOI
-
- Brokamp G, Valderrama N, Mittelbach M, Grandez CA, Barfod AS, Weigend M. Trade in palm products in North-Western South America. Bot Rev. 2011;77(4): 571–606. doi: 10.1007/s - DOI
-
- Eiserhardt WL, Pintaud J, Asmussen-lange C, Hahn J, Bernal R, Balslev H, et al.. Phytogeny and divergence times of Bactridinae (Arecaceae, Palmae) based on plastid and nuclear DNA sequences. Taxon. 2011;60(2): 485–498. doi: 10.1002/tax.602016 - DOI
-
- Baker WJ, Couvreur TLP. Global biogeography and diversification of palms sheds light on the evolution of tropical lineages. II. Diversification history and origin of regional assemblages. J Biogeogr. 2013;40(2): 286–298. doi: 10.1111/j.1365-2699.2012.02794.x - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

