Repression by the H-NS/YmoA histone-like protein complex enables IscR dependent regulation of the Yersinia T3SS
- PMID: 35901167
- PMCID: PMC9362927
- DOI: 10.1371/journal.pgen.1010321
Repression by the H-NS/YmoA histone-like protein complex enables IscR dependent regulation of the Yersinia T3SS
Abstract
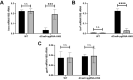
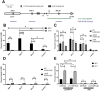
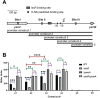
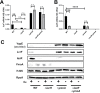
The type III secretion system (T3SS) is an appendage used by many bacterial pathogens, such as pathogenic Yersinia, to subvert host defenses. However, because the T3SS is energetically costly and immunogenic, it must be tightly regulated in response to environmental cues to enable survival in the host. Here we show that expression of the Yersinia Ysc T3SS master regulator, LcrF, is orchestrated by the opposing activities of the repressive H-NS/YmoA histone-like protein complex and induction by the iron and oxygen-regulated IscR transcription factor. While deletion of iscR or ymoA has been shown to decrease and increase LcrF expression and type III secretion, respectively, the role of H-NS in this system has not been definitively established because hns is an essential gene in Yersinia. Using CRISPRi knockdown of hns, we show that hns depletion causes derepression of lcrF. Furthermore, we find that while YmoA is dispensable for H-NS binding to the lcrF promoter, YmoA binding to H-NS is important for H-NS repressive activity. We bioinformatically identified three H-NS binding regions within the lcrF promoter and demonstrate binding of H-NS to these sites in vivo using chromatin immunoprecipitation. Using promoter truncation and binding site mutation analysis, we show that two of these H-NS binding regions are important for H-NS/YmoA-mediated repression of the lcrF promoter. Surprisingly, we find that IscR is dispensable for lcrF transcription in the absence of H-NS/YmoA. Indeed, IscR-dependent regulation of LcrF and type III secretion in response to changes in oxygen, such as those Yersinia is predicted to experience during host infection, only occurs in the presence of an H-NS/YmoA complex. These data suggest that, in the presence of host tissue cues that drive sufficient IscR expression, IscR can act as a roadblock to H-NS/YmoA-dependent repression of RNA polymerase at the lcrF promoter to turn on T3SS expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

