Constrained evolution of overlapping genes in viral host adaptation: Acquisition of glycosylation motifs in hepadnaviral precore/core genes
- PMID: 35901192
- PMCID: PMC9362955
- DOI: 10.1371/journal.ppat.1010739
Constrained evolution of overlapping genes in viral host adaptation: Acquisition of glycosylation motifs in hepadnaviral precore/core genes
Abstract
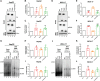
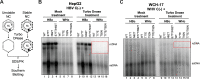
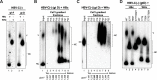
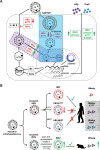
Hepadnaviruses use extensively overlapping genes to expand their coding capacity, especially the precore/core genes encode the precore and core proteins with mostly identical sequences but distinct functions. The precore protein of the woodchuck hepatitis virus (WHV) is N-glycosylated, in contrast to the precore of the human hepatitis B virus (HBV) that lacks N-glycosylation. To explore the roles of the N-linked glycosylation sites in precore and core functions, we substituted T77 and T92 in the WHV precore/core N-glycosylation motifs (75NIT77 and 90NDT92) with the corresponding HBV residues (E77 and N92) to eliminate the sequons. Conversely, these N-glycosylation sequons were introduced into the HBV precore/core gene by E77T and N92T substitutions. We found that N-glycosylation increased the levels of secreted precore gene products from both HBV and WHV. However, the HBV core (HBc) protein carrying the E77T substitution was defective in supporting virion secretion, and during infection, the HBc E77T and N92T substitutions impaired the formation of the covalently closed circular DNA (cccDNA), the critical viral DNA molecule responsible for establishing and maintaining infection. In cross-species complementation assays, both HBc and WHV core (WHc) proteins supported all steps of intracellular replication of the heterologous virus while WHc, with or without the N-glycosylation sequons, failed to interact with HBV envelope proteins for virion secretion. Interestingly, WHc supported more efficiently intracellular cccDNA amplification than HBc in the context of either HBV or WHV. These findings reveal novel determinants of precore secretion and core functions and illustrate strong constraints during viral host adaptation resulting from their compact genome and extensive use of overlapping genes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Characterization of Intracellular Precore-Derived Proteins and Their Functions in Hepatitis B Virus-Infected Human Hepatocytes.mBio. 2023 Feb 28;14(1):e0350122. doi: 10.1128/mbio.03501-22. Epub 2023 Jan 30. mBio. 2023. PMID: 36715515 Free PMC article.
-
Characterization and Application of Precore/Core-Related Antigens in Animal Models of Hepatitis B Virus Infection.Hepatology. 2021 Jul;74(1):99-115. doi: 10.1002/hep.31720. Hepatology. 2021. PMID: 33458844 Free PMC article.
-
Conserved Lysine Residues of Hepatitis B Virus Core Protein Are Not Required for Covalently Closed Circular DNA Formation.J Virol. 2022 Aug 10;96(15):e0071822. doi: 10.1128/jvi.00718-22. Epub 2022 Jul 18. J Virol. 2022. PMID: 35867543 Free PMC article.
-
Tracing the evolutionary history of hepadnaviruses in terms of e antigen and middle envelope protein expression or processing.Virus Res. 2020 Jan 15;276:197825. doi: 10.1016/j.virusres.2019.197825. Epub 2019 Nov 27. Virus Res. 2020. PMID: 31785305 Free PMC article. Review.
-
Silencing hepatitis B virus covalently closed circular DNA: The potential of an epigenetic therapy approach.World J Gastroenterol. 2021 Jun 21;27(23):3182-3207. doi: 10.3748/wjg.v27.i23.3182. World J Gastroenterol. 2021. PMID: 34163105 Free PMC article. Review.
Cited by
-
Hepatitis B Virus Capsid: The Core in Productive Entry and Covalently Closed Circular DNA Formation.Viruses. 2023 Feb 28;15(3):642. doi: 10.3390/v15030642. Viruses. 2023. PMID: 36992351 Free PMC article. Review.
-
Deviated binding of anti-HBV nucleoside analog E-CFCP-TP to the reverse transcriptase active site attenuates the effect of drug-resistant mutations.Sci Rep. 2024 Jul 8;14(1):15742. doi: 10.1038/s41598-024-66505-z. Sci Rep. 2024. PMID: 38977798 Free PMC article.
-
Detection of Hepatitis B Virus Covalently Closed Circular DNA and Intermediates in Its Formation.Methods Mol Biol. 2024;2837:99-111. doi: 10.1007/978-1-0716-4027-2_9. Methods Mol Biol. 2024. PMID: 39044078 Free PMC article.
-
Characterization of Intracellular Precore-Derived Proteins and Their Functions in Hepatitis B Virus-Infected Human Hepatocytes.mBio. 2023 Feb 28;14(1):e0350122. doi: 10.1128/mbio.03501-22. Epub 2023 Jan 30. mBio. 2023. PMID: 36715515 Free PMC article.
References
-
- WHO. Global hepatitis report 2017: World Health Organization; 2017.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

