Natural Evolution Provides Strong Hints about Laboratory Evolution of Designer Enzymes
- PMID: 35901204
- PMCID: PMC9351539
- DOI: 10.1073/pnas.2207904119
Natural Evolution Provides Strong Hints about Laboratory Evolution of Designer Enzymes
Abstract
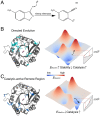
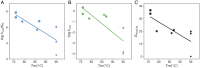
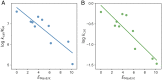
Laboratory evolution combined with computational enzyme design provides the opportunity to generate novel biocatalysts. Nevertheless, it has been challenging to understand how laboratory evolution optimizes designer enzymes by introducing seemingly random mutations. A typical enzyme optimized with laboratory evolution is the abiological Kemp eliminase, initially designed by grafting active site residues into a natural protein scaffold. Here, we relate the catalytic power of laboratory-evolved Kemp eliminases to the statistical energy ([Formula: see text]) inferred from their natural homologous sequences using the maximum entropy model. The [Formula: see text] of designs generated by directed evolution is correlated with enhanced activity and reduced stability, thus displaying a stability-activity trade-off. In contrast, the [Formula: see text] for mutants in catalytic-active remote regions (in which remote residues are important for catalysis) is strongly anticorrelated with the activity. These findings provide an insight into the role of protein scaffolds in the adaption to new enzymatic functions. It also indicates that the valley in the [Formula: see text] landscape can guide enzyme design for abiological catalysis. Overall, the connection between laboratory and natural evolution contributes to understanding what is optimized in the laboratory and how new enzymatic function emerges in nature, and provides guidance for computational enzyme design.
Keywords: designer enzyme; directed evolution; enzyme architecture; enzyme design; natural evolution.
Conflict of interest statement
The authors declare no competing interest.
Figures

Similar articles
-
Evolutionary optimization of computationally designed enzymes: Kemp eliminases of the KE07 series.J Mol Biol. 2010 Mar 5;396(4):1025-42. doi: 10.1016/j.jmb.2009.12.031. Epub 2009 Dec 28. J Mol Biol. 2010. PMID: 20036254
-
The role of reorganization energy in rational enzyme design.Curr Opin Chem Biol. 2014 Aug;21:34-41. doi: 10.1016/j.cbpa.2014.03.011. Epub 2014 Apr 24. Curr Opin Chem Biol. 2014. PMID: 24769299 Review.
-
Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.Proc Natl Acad Sci U S A. 2012 Jun 26;109(26):10358-63. doi: 10.1073/pnas.1121063109. Epub 2012 Jun 8. Proc Natl Acad Sci U S A. 2012. PMID: 22685214 Free PMC article.
-
Enriching productive mutational paths accelerates enzyme evolution.Nat Chem Biol. 2024 Dec;20(12):1662-1669. doi: 10.1038/s41589-024-01712-3. Epub 2024 Sep 11. Nat Chem Biol. 2024. PMID: 39261644 Free PMC article.
-
Designing better enzymes: Insights from directed evolution.Curr Opin Struct Biol. 2021 Apr;67:212-218. doi: 10.1016/j.sbi.2020.12.015. Epub 2021 Jan 29. Curr Opin Struct Biol. 2021. PMID: 33517098 Review.
Cited by
-
Harnessing generative AI to decode enzyme catalysis and evolution for enhanced engineering.Natl Sci Rev. 2023 Dec 28;10(12):nwad331. doi: 10.1093/nsr/nwad331. eCollection 2023 Dec. Natl Sci Rev. 2023. PMID: 38299119 Free PMC article. Review.
-
EnzyHTP Computational Directed Evolution with Adaptive Resource Allocation.J Chem Inf Model. 2023 Sep 11;63(17):5650-5659. doi: 10.1021/acs.jcim.3c00618. Epub 2023 Aug 23. J Chem Inf Model. 2023. PMID: 37611241 Free PMC article.
-
SubTuner leverages physics-based modeling to complement AI in enzyme engineering toward non-native substrates.Chem Catal. 2025 Jun 19;5(6):101334. doi: 10.1016/j.checat.2025.101334. Epub 2025 Mar 28. Chem Catal. 2025. PMID: 40717709
-
Harnessing Generative AI to Decode Enzyme Catalysis and Evolution for Enhanced Engineering.bioRxiv [Preprint]. 2023 Oct 12:2023.10.10.561808. doi: 10.1101/2023.10.10.561808. bioRxiv. 2023. Update in: Natl Sci Rev. 2023 Dec 28;10(12):nwad331. doi: 10.1093/nsr/nwad331. PMID: 37873334 Free PMC article. Updated. Preprint.
-
Substrate Positioning Dynamics Involves a Non-Electrostatic Component to Mediate Catalysis.J Phys Chem Lett. 2023 Dec 21;14(50):11480-11489. doi: 10.1021/acs.jpclett.3c02444. Epub 2023 Dec 12. J Phys Chem Lett. 2023. PMID: 38085952 Free PMC article.
References
-
- Davidi D., Longo L. M., Jabłońska J., Milo R., Tawfik D. S., A bird’s-eye view of enzyme evolution: Chemical, physicochemical, and physiological considerations. Chem. Rev. 118, 8786–8797 (2018). - PubMed
-
- Röthlisberger D., et al. , Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

