Protease-activated receptors in health and disease
- PMID: 35901239
- PMCID: PMC9662810
- DOI: 10.1152/physrev.00044.2021
Protease-activated receptors in health and disease
Abstract
Proteases are signaling molecules that specifically control cellular functions by cleaving protease-activated receptors (PARs). The four known PARs are members of the large family of G protein-coupled receptors. These transmembrane receptors control most physiological and pathological processes and are the target of a large proportion of therapeutic drugs. Signaling proteases include enzymes from the circulation; from immune, inflammatory epithelial, and cancer cells; as well as from commensal and pathogenic bacteria. Advances in our understanding of the structure and function of PARs provide insights into how diverse proteases activate these receptors to regulate physiological and pathological processes in most tissues and organ systems. The realization that proteases and PARs are key mediators of disease, coupled with advances in understanding the atomic level structure of PARs and their mechanisms of signaling in subcellular microdomains, has spurred the development of antagonists, some of which have advanced to the clinic. Herein we review the discovery, structure, and function of this receptor system, highlight the contribution of PARs to homeostatic control, and discuss the potential of PAR antagonists for the treatment of major diseases.
Keywords: antagonists; disease; homeostasis; proteases; receptors.
Conflict of interest statement
N. W. Bunnett is a founding scientist of Endosome Therapeutics Inc. None of the other authors has any conflicts of interest, financial or otherwise, to disclose.
Figures



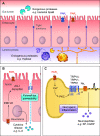
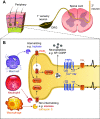

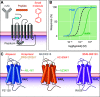
References
-
- Schmidt A. Neue Untersuchungen ueber die Fasserstoffesgerinnung. Pflüger Arch 6: 413–538, 1872. doi: 10.1007/BF01612263. - DOI
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

