Impaired ketogenesis ties metabolism to T cell dysfunction in COVID-19
- PMID: 35901960
- PMCID: PMC9428867
- DOI: 10.1038/s41586-022-05128-8
Impaired ketogenesis ties metabolism to T cell dysfunction in COVID-19
Abstract
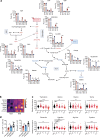
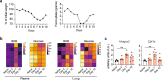
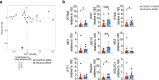
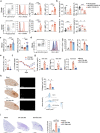
Anorexia and fasting are host adaptations to acute infection, and induce a metabolic switch towards ketogenesis and the production of ketone bodies, including β-hydroxybutyrate (BHB)1-6. However, whether ketogenesis metabolically influences the immune response in pulmonary infections remains unclear. Here we show that the production of BHB is impaired in individuals with SARS-CoV-2-induced acute respiratory distress syndrome (ARDS) but not in those with influenza-induced ARDS. We found that BHB promotes both the survival of and the production of interferon-γ by CD4+ T cells. Applying a metabolic-tracing analysis, we established that BHB provides an alternative carbon source to fuel oxidative phosphorylation (OXPHOS) and the production of bioenergetic amino acids and glutathione, which is important for maintaining the redox balance. T cells from patients with SARS-CoV-2-induced ARDS were exhausted and skewed towards glycolysis, but could be metabolically reprogrammed by BHB to perform OXPHOS, thereby increasing their functionality. Finally, we show in mice that a ketogenic diet and the delivery of BHB as a ketone ester drink restores CD4+ T cell metabolism and function in severe respiratory infections, ultimately reducing the mortality of mice infected with SARS-CoV-2. Altogether, our data reveal that BHB is an alternative source of carbon that promotes T cell responses in pulmonary viral infections, and highlight impaired ketogenesis as a potential confounding factor in severe COVID-19.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures









Comment in
-
Fasting as key tone for COVID immunity.Nat Metab. 2022 Oct;4(10):1229-1231. doi: 10.1038/s42255-022-00646-1. Nat Metab. 2022. PMID: 36180792 No abstract available.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

